Anwai Archit
BioimageAIpub: a toolbox for AI-ready bioimaging data publishing
Dec 17, 2025Abstract:Modern bioimage analysis approaches are data hungry, making it necessary for researchers to scavenge data beyond those collected within their (bio)imaging facilities. In addition to scale, bioimaging datasets must be accompanied with suitable, high-quality annotations and metadata. Although established data repositories such as the Image Data Resource (IDR) and BioImage Archive offer rich metadata, their contents typically cannot be directly consumed by image analysis tools without substantial data wrangling. Such a tedious assembly and conversion of (meta)data can account for a dedicated amount of time investment for researchers, hindering the development of more powerful analysis tools. Here, we introduce BioimageAIpub, a workflow that streamlines bioimaging data conversion, enabling a seamless upload to HuggingFace, a widely used platform for sharing machine learning datasets and models.
Tiling artifacts and trade-offs of feature normalization in the segmentation of large biological images
Mar 25, 2025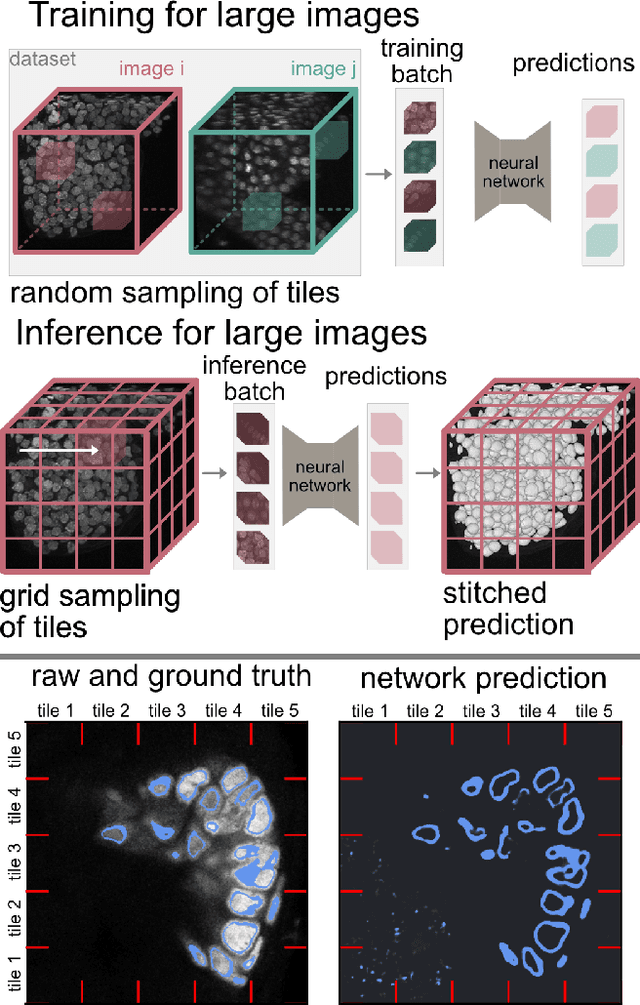
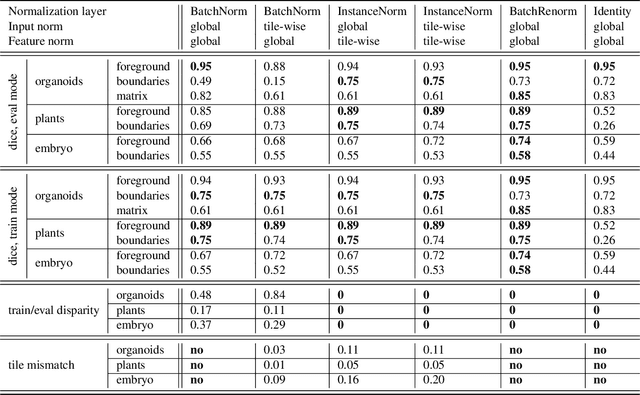
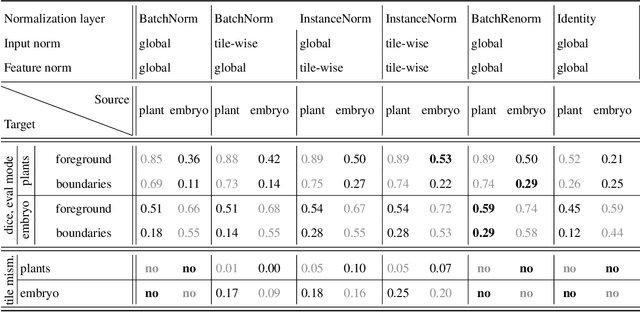
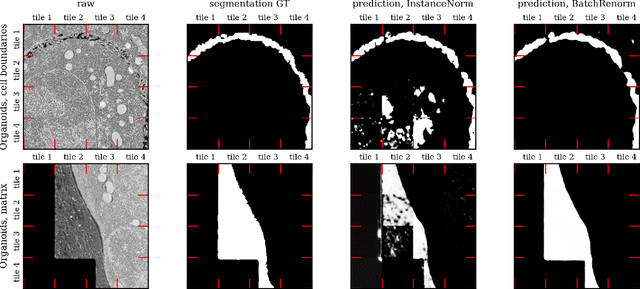
Abstract:Segmentation of very large images is a common problem in microscopy, medical imaging or remote sensing. The problem is usually addressed by sliding window inference, which can theoretically lead to seamlessly stitched predictions. However, in practice many of the popular pipelines still suffer from tiling artifacts. We investigate the root cause of these issues and show that they stem from the normalization layers within the neural networks. We propose indicators to detect normalization issues and further explore the trade-offs between artifact-free and high-quality predictions, using three diverse microscopy datasets as examples. Finally, we propose to use BatchRenorm as the most suitable normalization strategy, which effectively removes tiling artifacts and enhances transfer performance, thereby improving the reusability of trained networks for new datasets.
MedicoSAM: Towards foundation models for medical image segmentation
Jan 20, 2025Abstract:Medical image segmentation is an important analysis task in clinical practice and research. Deep learning has massively advanced the field, but current approaches are mostly based on models trained for a specific task. Training such models or adapting them to a new condition is costly due to the need for (manually) labeled data. The emergence of vision foundation models, especially Segment Anything, offers a path to universal segmentation for medical images, overcoming these issues. Here, we study how to improve Segment Anything for medical images by comparing different finetuning strategies on a large and diverse dataset. We evaluate the finetuned models on a wide range of interactive and (automatic) semantic segmentation tasks. We find that the performance can be clearly improved for interactive segmentation. However, semantic segmentation does not benefit from pretraining on medical images. Our best model, MedicoSAM, is publicly available at https://github.com/computational-cell-analytics/medico-sam. We show that it is compatible with existing tools for data annotation and believe that it will be of great practical value.
ViM-UNet: Vision Mamba for Biomedical Segmentation
Apr 11, 2024Abstract:CNNs, most notably the UNet, are the default architecture for biomedical segmentation. Transformer-based approaches, such as UNETR, have been proposed to replace them, benefiting from a global field of view, but suffering from larger runtimes and higher parameter counts. The recent Vision Mamba architecture offers a compelling alternative to transformers, also providing a global field of view, but at higher efficiency. Here, we introduce ViM-UNet, a novel segmentation architecture based on it and compare it to UNet and UNETR for two challenging microscopy instance segmentation tasks. We find that it performs similarly or better than UNet, depending on the task, and outperforms UNETR while being more efficient. Our code is open source and documented at https://github.com/constantinpape/torch-em/blob/main/vimunet.md.
Probabilistic Domain Adaptation for Biomedical Image Segmentation
Mar 21, 2023Abstract:Segmentation is a key analysis tasks in biomedical imaging. Given the many different experimental settings in this field, the lack of generalization limits the use of deep learning in practice. Domain adaptation is a promising remedy: it trains a model for a given task on a source dataset with labels and adapts it to a target dataset without additional labels. We introduce a probabilistic domain adaptation method, building on self-training approaches and the Probabilistic UNet. We use the latter to sample multiple segmentation hypothesis to implement better pseudo-label filtering. We further study joint and separate source-target training strategies and evaluate our method on three challenging domain adaptation tasks for biomedical segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge