Anusri Pampari
DART-Eval: A Comprehensive DNA Language Model Evaluation Benchmark on Regulatory DNA
Dec 06, 2024



Abstract:Recent advances in self-supervised models for natural language, vision, and protein sequences have inspired the development of large genomic DNA language models (DNALMs). These models aim to learn generalizable representations of diverse DNA elements, potentially enabling various genomic prediction, interpretation and design tasks. Despite their potential, existing benchmarks do not adequately assess the capabilities of DNALMs on key downstream applications involving an important class of non-coding DNA elements critical for regulating gene activity. In this study, we introduce DART-Eval, a suite of representative benchmarks specifically focused on regulatory DNA to evaluate model performance across zero-shot, probed, and fine-tuned scenarios against contemporary ab initio models as baselines. Our benchmarks target biologically meaningful downstream tasks such as functional sequence feature discovery, predicting cell-type specific regulatory activity, and counterfactual prediction of the impacts of genetic variants. We find that current DNALMs exhibit inconsistent performance and do not offer compelling gains over alternative baseline models for most tasks, while requiring significantly more computational resources. We discuss potentially promising modeling, data curation, and evaluation strategies for the next generation of DNALMs. Our code is available at https://github.com/kundajelab/DART-Eval.
Unsupervised Calibration under Covariate Shift
Jun 29, 2020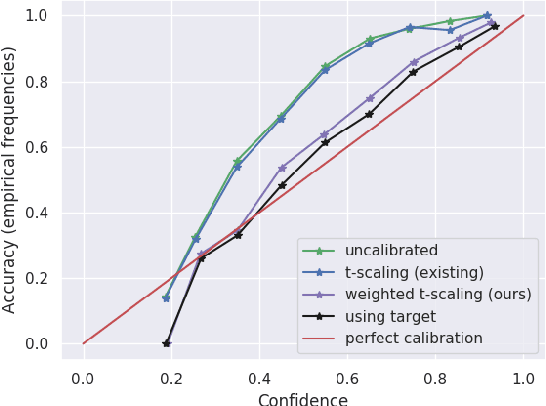
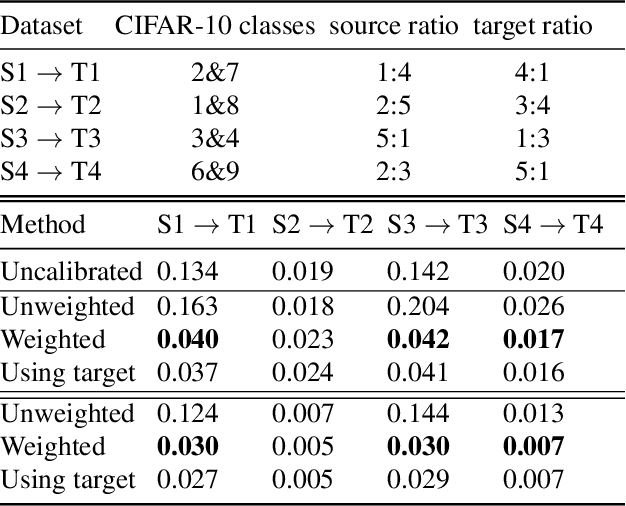
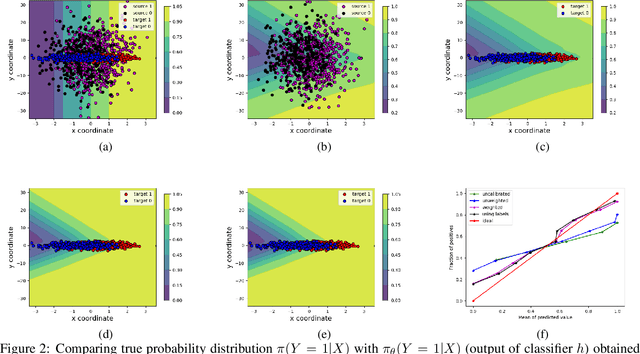
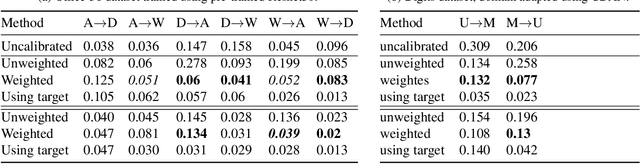
Abstract:A probabilistic model is said to be calibrated if its predicted probabilities match the corresponding empirical frequencies. Calibration is important for uncertainty quantification and decision making in safety-critical applications. While calibration of classifiers has been widely studied, we find that calibration is brittle and can be easily lost under minimal covariate shifts. Existing techniques, including domain adaptation ones, primarily focus on prediction accuracy and do not guarantee calibration neither in theory nor in practice. In this work, we formally introduce the problem of calibration under domain shift, and propose an importance sampling based approach to address it. We evaluate and discuss the efficacy of our method on both real-world datasets and synthetic datasets.
emrQA: A Large Corpus for Question Answering on Electronic Medical Records
Sep 03, 2018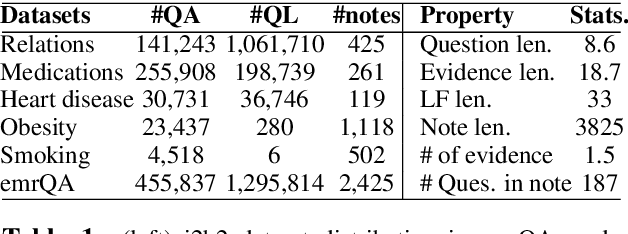
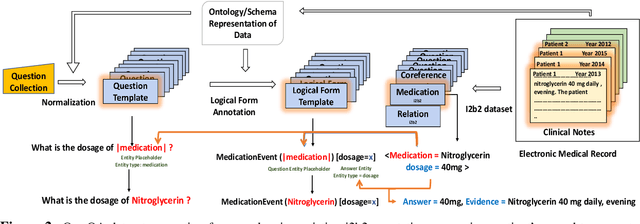
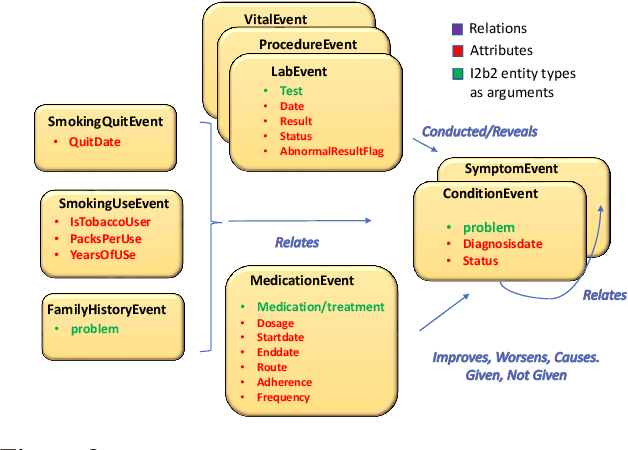
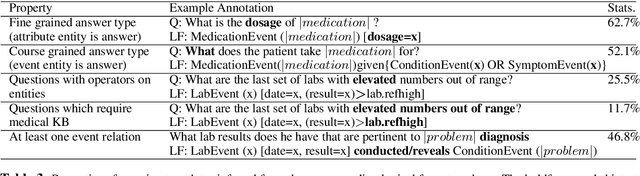
Abstract:We propose a novel methodology to generate domain-specific large-scale question answering (QA) datasets by re-purposing existing annotations for other NLP tasks. We demonstrate an instance of this methodology in generating a large-scale QA dataset for electronic medical records by leveraging existing expert annotations on clinical notes for various NLP tasks from the community shared i2b2 datasets. The resulting corpus (emrQA) has 1 million question-logical form and 400,000+ question-answer evidence pairs. We characterize the dataset and explore its learning potential by training baseline models for question to logical form and question to answer mapping.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge