Anton Nikolaev
MorphoSeg: An Uncertainty-Aware Deep Learning Method for Biomedical Segmentation of Complex Cellular Morphologies
Sep 25, 2024



Abstract:Deep learning has revolutionized medical and biological imaging, particularly in segmentation tasks. However, segmenting biological cells remains challenging due to the high variability and complexity of cell shapes. Addressing this challenge requires high-quality datasets that accurately represent the diverse morphologies found in biological cells. Existing cell segmentation datasets are often limited by their focus on regular and uniform shapes. In this paper, we introduce a novel benchmark dataset of Ntera-2 (NT2) cells, a pluripotent carcinoma cell line, exhibiting diverse morphologies across multiple stages of differentiation, capturing the intricate and heterogeneous cellular structures that complicate segmentation tasks. To address these challenges, we propose an uncertainty-aware deep learning framework for complex cellular morphology segmentation (MorphoSeg) by incorporating sampling of virtual outliers from low-likelihood regions during training. Our comprehensive experimental evaluations against state-of-the-art baselines demonstrate that MorphoSeg significantly enhances segmentation accuracy, achieving up to a 7.74% increase in the Dice Similarity Coefficient (DSC) and a 28.36% reduction in the Hausdorff Distance. These findings highlight the effectiveness of our dataset and methodology in advancing cell segmentation capabilities, especially for complex and variable cell morphologies. The dataset and source code is publicly available at https://github.com/RanchoGoose/MorphoSeg.
FishNet: Deep Neural Networks for Low-Cost Fish Stock Estimation
Mar 16, 2024

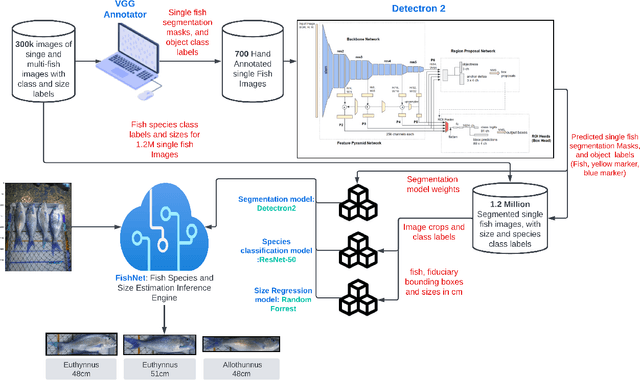
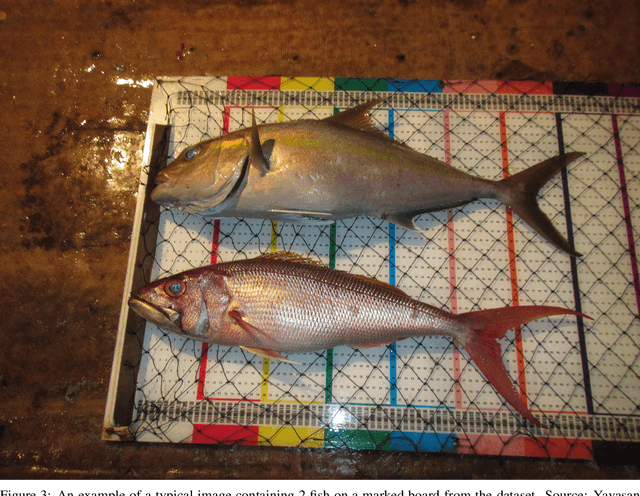
Abstract:Fish stock assessment often involves manual fish counting by taxonomy specialists, which is both time-consuming and costly. We propose an automated computer vision system that performs both taxonomic classification and fish size estimation from images taken with a low-cost digital camera. The system first performs object detection and segmentation using a Mask R-CNN to identify individual fish from images containing multiple fish, possibly consisting of different species. Then each fish species is classified and the predicted length using separate machine learning models. These models are trained on a dataset of 50,000 hand-annotated images containing 163 different fish species, ranging in length from 10cm to 250cm. Evaluated on held-out test data, our system achieves a $92\%$ intersection over union on the fish segmentation task, a $89\%$ top-1 classification accuracy on single fish species classification, and a $2.3$~cm mean error on the fish length estimation task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge