Antoine Pirovano
Improving Interpretability for Computer-aided Diagnosis tools on Whole Slide Imaging with Multiple Instance Learning and Gradient-based Explanations
Sep 29, 2020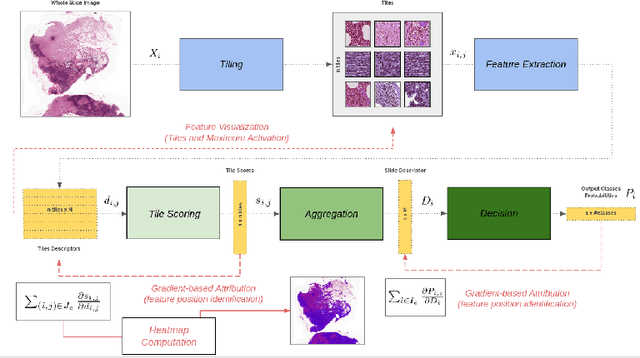
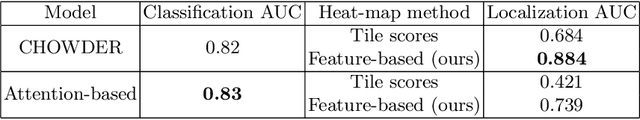
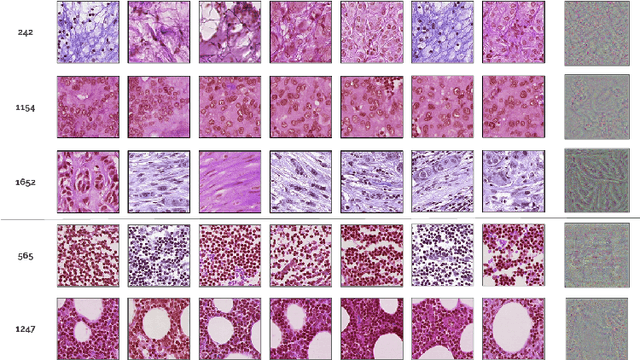
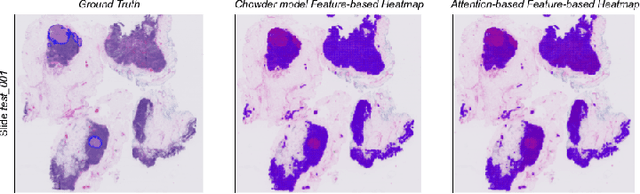
Abstract:Deep learning methods are widely used for medical applications to assist medical doctors in their daily routines. While performances reach expert's level, interpretability (highlight how and what a trained model learned and why it makes a specific decision) is the next important challenge that deep learning methods need to answer to be fully integrated in the medical field. In this paper, we address the question of interpretability in the context of whole slide images (WSI) classification. We formalize the design of WSI classification architectures and propose a piece-wise interpretability approach, relying on gradient-based methods, feature visualization and multiple instance learning context. We aim at explaining how the decision is made based on tile level scoring, how these tile scores are decided and which features are used and relevant for the task. After training two WSI classification architectures on Camelyon-16 WSI dataset, highlighting discriminative features learned, and validating our approach with pathologists, we propose a novel manner of computing interpretability slide-level heat-maps, based on the extracted features, that improves tile-level classification performances by more than 29% for AUC.
Regression Constraint for an Explainable Cervical Cancer Classifier
Aug 19, 2019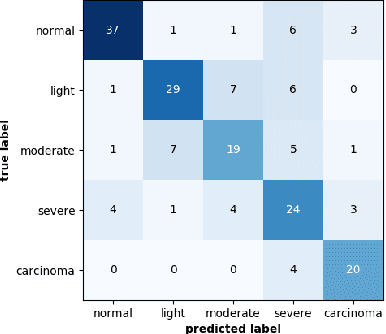
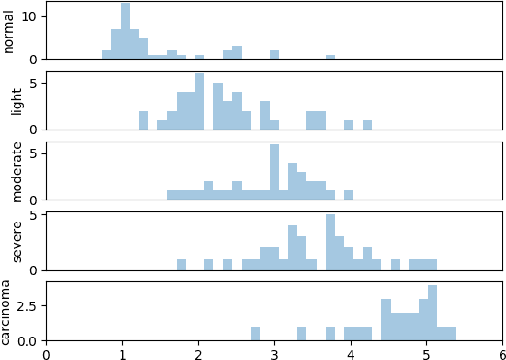
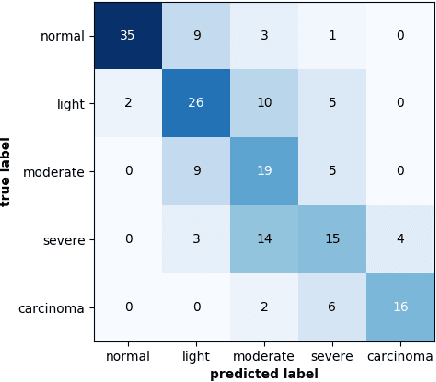
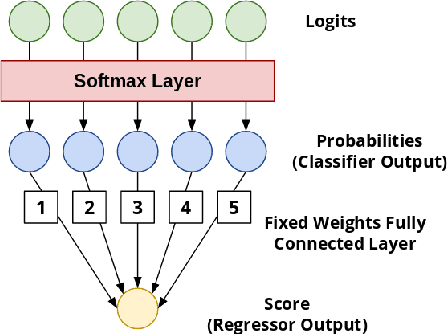
Abstract:This article adresses the problem of automatic squamous cells classification for cervical cancer screening using Deep Learning methods. We study different architectures on a public dataset called Herlev dataset, which consists in classifying cells, obtained by cervical pap smear, regarding the severity of the abnormalities they represent. Furthermore, we use an attribution method to understand which cytomorphological features are actually learned as discriminative to classify severity of the abnormalities. Through this paper, we show how we trained a performant classifier: 74.5\% accuracy on severity classification and 94\% accuracy on normal/abnormal classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge