Annamaria Porreca
Enriched Functional Tree-Based Classifiers: A Novel Approach Leveraging Derivatives and Geometric Features
Sep 26, 2024



Abstract:The positioning of this research falls within the scalar-on-function classification literature, a field of significant interest across various domains, particularly in statistics, mathematics, and computer science. This study introduces an advanced methodology for supervised classification by integrating Functional Data Analysis (FDA) with tree-based ensemble techniques for classifying high-dimensional time series. The proposed framework, Enriched Functional Tree-Based Classifiers (EFTCs), leverages derivative and geometric features, benefiting from the diversity inherent in ensemble methods to further enhance predictive performance and reduce variance. While our approach has been tested on the enrichment of Functional Classification Trees (FCTs), Functional K-NN (FKNN), Functional Random Forest (FRF), Functional XGBoost (FXGB), and Functional LightGBM (FLGBM), it could be extended to other tree-based and non-tree-based classifiers, with appropriate considerations emerging from this investigation. Through extensive experimental evaluations on seven real-world datasets and six simulated scenarios, this proposal demonstrates fascinating improvements over traditional approaches, providing new insights into the application of FDA in complex, high-dimensional learning problems.
Augmented Functional Random Forests: Classifier Construction and Unbiased Functional Principal Components Importance through Ad-Hoc Conditional Permutations
Aug 23, 2024



Abstract:This paper introduces a novel supervised classification strategy that integrates functional data analysis (FDA) with tree-based methods, addressing the challenges of high-dimensional data and enhancing the classification performance of existing functional classifiers. Specifically, we propose augmented versions of functional classification trees and functional random forests, incorporating a new tool for assessing the importance of functional principal components. This tool provides an ad-hoc method for determining unbiased permutation feature importance in functional data, particularly when dealing with correlated features derived from successive derivatives. Our study demonstrates that these additional features can significantly enhance the predictive power of functional classifiers. Experimental evaluations on both real-world and simulated datasets showcase the effectiveness of the proposed methodology, yielding promising results compared to existing methods.
Demystifying Functional Random Forests: Novel Explainability Tools for Model Transparency in High-Dimensional Spaces
Aug 22, 2024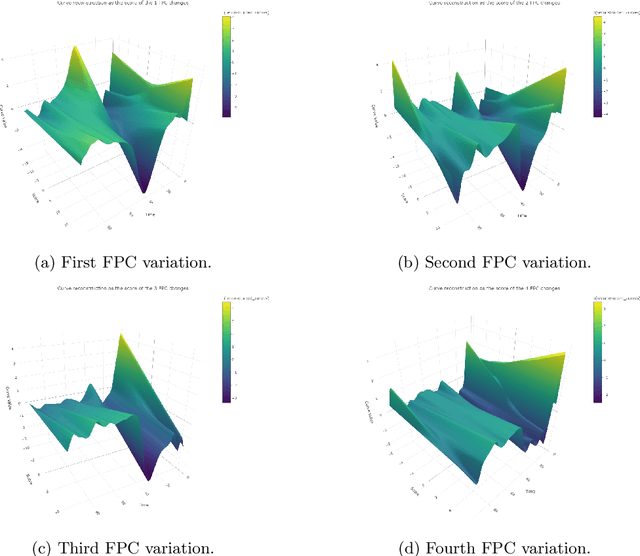
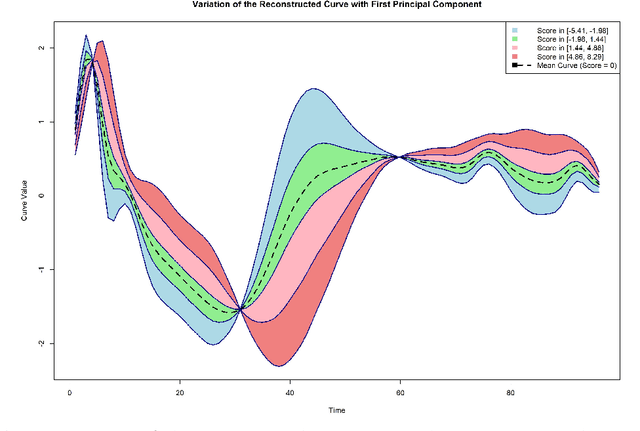
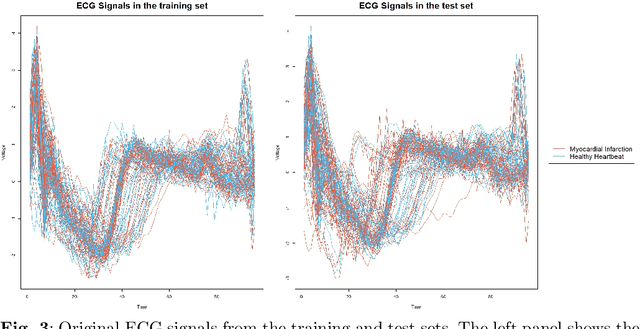
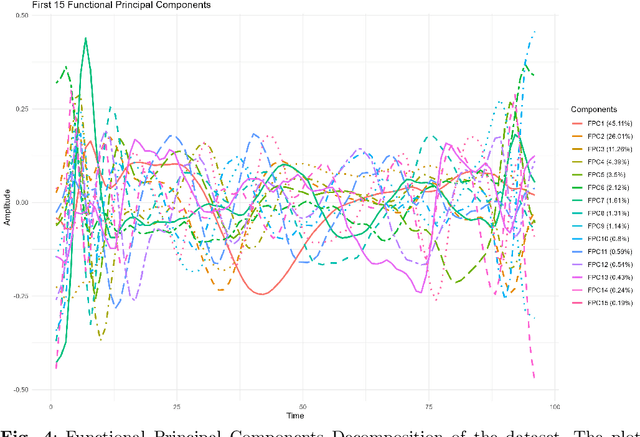
Abstract:The advent of big data has raised significant challenges in analysing high-dimensional datasets across various domains such as medicine, ecology, and economics. Functional Data Analysis (FDA) has proven to be a robust framework for addressing these challenges, enabling the transformation of high-dimensional data into functional forms that capture intricate temporal and spatial patterns. However, despite advancements in functional classification methods and very high performance demonstrated by combining FDA and ensemble methods, a critical gap persists in the literature concerning the transparency and interpretability of black-box models, e.g. Functional Random Forests (FRF). In response to this need, this paper introduces a novel suite of explainability tools to illuminate the inner mechanisms of FRF. We propose using Functional Partial Dependence Plots (FPDPs), Functional Principal Component (FPC) Probability Heatmaps, various model-specific and model-agnostic FPCs' importance metrics, and the FPC Internal-External Importance and Explained Variance Bubble Plot. These tools collectively enhance the transparency of FRF models by providing a detailed analysis of how individual FPCs contribute to model predictions. By applying these methods to an ECG dataset, we demonstrate the effectiveness of these tools in revealing critical patterns and improving the explainability of FRF.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge