Andrew R. Cohen
Dept Electri. Comput. Eng., Drexel Univ.
The cell signaling structure function
Jan 11, 2024Abstract:Live cell microscopy captures 5-D $(x,y,z,channel,time)$ movies that display patterns of cellular motion and signaling dynamics. We present here an approach to finding spatiotemporal patterns of cell signaling dynamics in 5-D live cell microscopy movies unique in requiring no a priori knowledge of expected pattern dynamics, and no training data. The proposed cell signaling structure function (SSF) is a Kolmogorov structure function that optimally measures cell signaling state as nuclear intensity w.r.t. surrounding cytoplasm, a significant improvement compared to the current state-of-the-art cytonuclear ratio. SSF kymographs store at each spatiotemporal cell centroid the SSF value, or a functional output such as velocity. Patterns of similarity are identified via the metric normalized compression distance (NCD). The NCD is a reproducing kernel for a Hilbert space that represents the input SSF kymographs as points in a low dimensional embedding that optimally captures the pattern similarity identified by the NCD throughout the space. The only parameter is the expected cell radii ($\mu m$). A new formulation of the cluster structure function optimally estimates how meaningful an embedding from the RKHS representation. Results are presented quantifying the impact of ERK and AKT signaling between different oncogenic mutations, and by the relation between ERK signaling and cellular velocity patterns for movies of 2-D monolayers of human breast epithelial (MCF10A) cells, 3-D MCF10A spheroids under optogenetic manipulation of ERK, and human induced pluripotent stem cells .
The cluster structure function
Jan 11, 2022
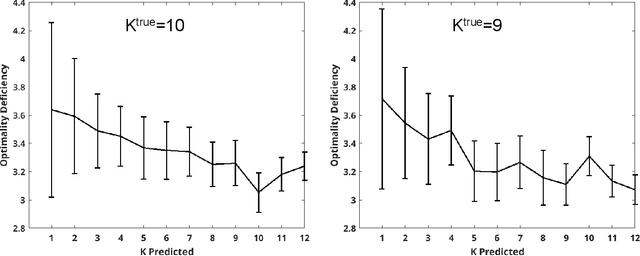

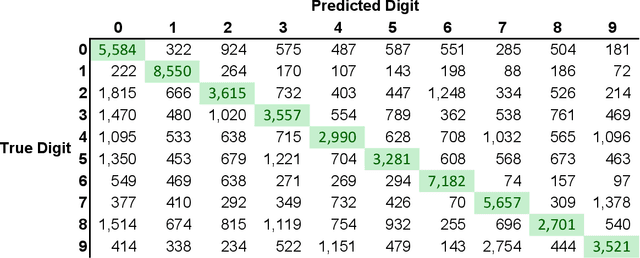
Abstract:For each partition of a data set into a given number of parts there is a partition such that every part is as much as possible a good model (an "algorithmic sufficient statistic") for the data in that part. Since this can be done for every number between one and the number of data, the result is a function, the cluster structure function. It maps the number of parts of a partition to values related to the deficiencies of being good models by the parts. Such a function starts with a value at least zero for no partition of the data set and descents to zero for the partition of the data set into singleton parts. The optimal clustering is the one chosen to minimize the cluster structure function. The theory behind the method is expressed in algorithmic information theory (Kolmogorov complexity). In practice the Kolmogorov complexities involved are approximated by a concrete compressor. We give examples using real data sets: the MNIST handwritten digits and the segmentation of real cells as used in stem cell research.
Web Similarity
Feb 20, 2015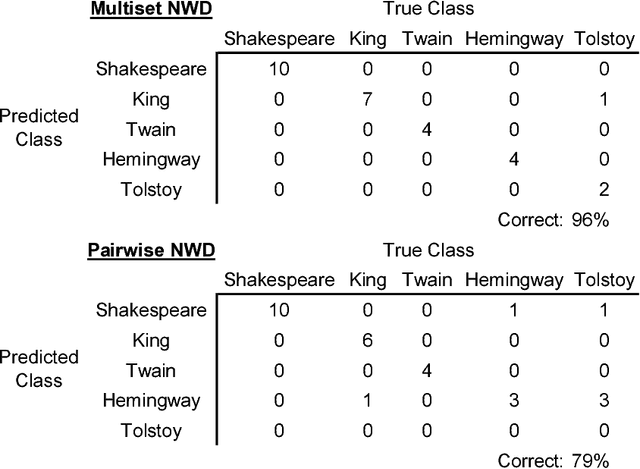
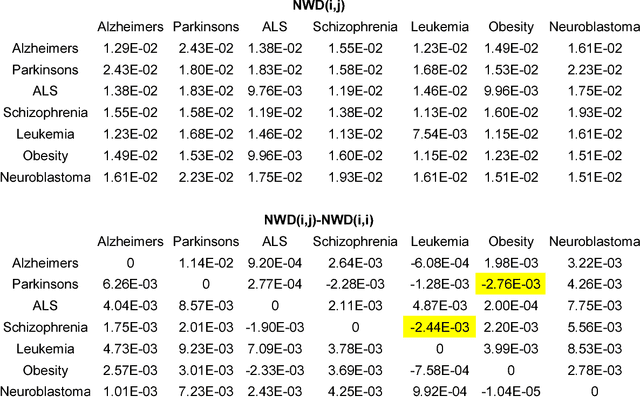
Abstract:Normalized web distance (NWD) is a similarity or normalized semantic distance based on the World Wide Web or any other large electronic database, for instance Wikipedia, and a search engine that returns reliable aggregate page counts. For sets of search terms the NWD gives a similarity on a scale from 0 (identical) to 1 (completely different). The NWD approximates the similarity according to all (upper semi)computable properties. We develop the theory and give applications. The derivation of the NWD method is based on Kolmogorov complexity.
Normalized Google Distance of Multisets with Applications
Aug 14, 2013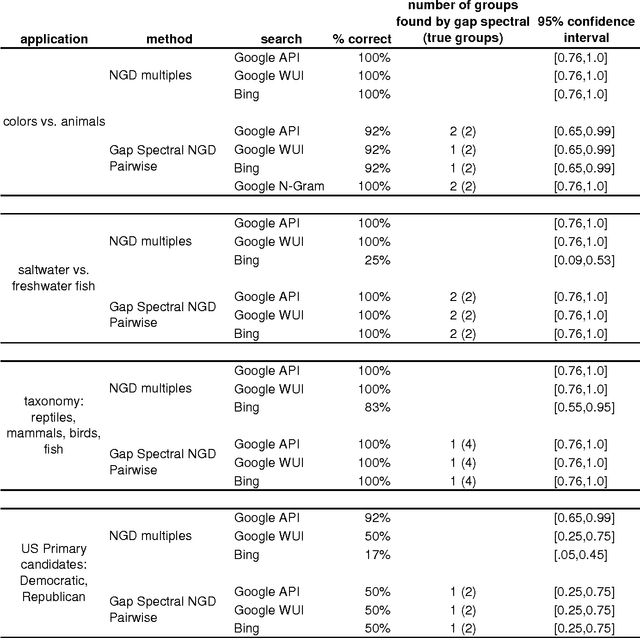
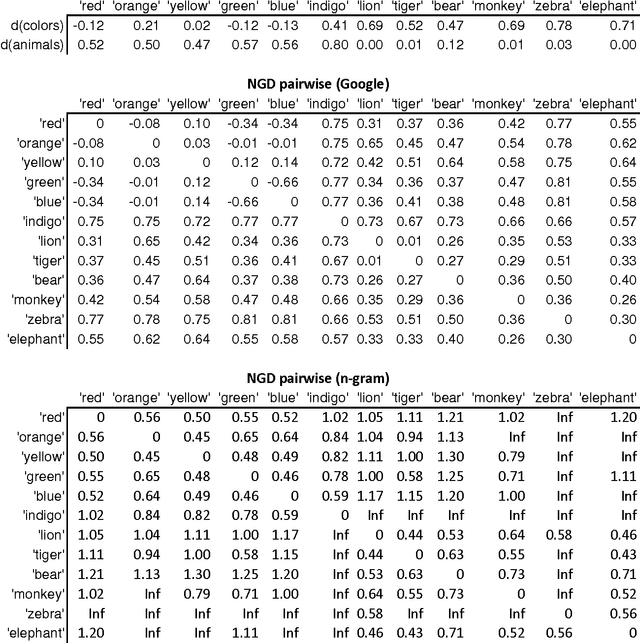
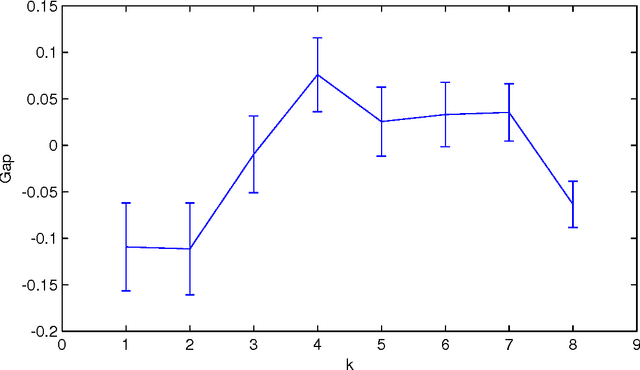
Abstract:Normalized Google distance (NGD) is a relative semantic distance based on the World Wide Web (or any other large electronic database, for instance Wikipedia) and a search engine that returns aggregate page counts. The earlier NGD between pairs of search terms (including phrases) is not sufficient for all applications. We propose an NGD of finite multisets of search terms that is better for many applications. This gives a relative semantics shared by a multiset of search terms. We give applications and compare the results with those obtained using the pairwise NGD. The derivation of NGD method is based on Kolmogorov complexity.
Normalized Compression Distance of Multisets with Applications
Mar 29, 2013

Abstract:Normalized compression distance (NCD) is a parameter-free, feature-free, alignment-free, similarity measure between a pair of finite objects based on compression. However, it is not sufficient for all applications. We propose an NCD of finite multisets (a.k.a. multiples) of finite objects that is also a metric. Previously, attempts to obtain such an NCD failed. We cover the entire trajectory from theoretical underpinning to feasible practice. The new NCD for multisets is applied to retinal progenitor cell classification questions and to related synthetically generated data that were earlier treated with the pairwise NCD. With the new method we achieved significantly better results. Similarly for questions about axonal organelle transport. We also applied the new NCD to handwritten digit recognition and improved classification accuracy significantly over that of pairwise NCD by incorporating both the pairwise and NCD for multisets. In the analysis we use the incomputable Kolmogorov complexity that for practical purposes is approximated from above by the length of the compressed version of the file involved, using a real-world compression program. Index Terms--- Normalized compression distance, multisets or multiples, pattern recognition, data mining, similarity, classification, Kolmogorov complexity, retinal progenitor cells, synthetic data, organelle transport, handwritten character recognition
* LaTeX 28 pages, 3 figures. This version is changed from the preliminary version to the final version. Updates of the theory. How to compute it, special recepies for classification, more applications and better results (see abstract and especially the detailed results in the paper). The title was changed to reflect this. In v4 corrected the proof of Theorem III-7
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge