Andrew P. Norgan
Performance of externally validated machine learning models based on histopathology images for the diagnosis, classification, prognosis, or treatment outcome prediction in female breast cancer: A systematic review
Dec 09, 2023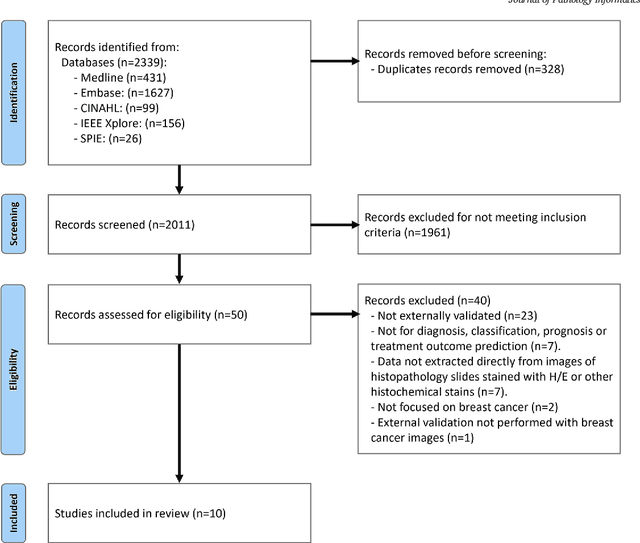
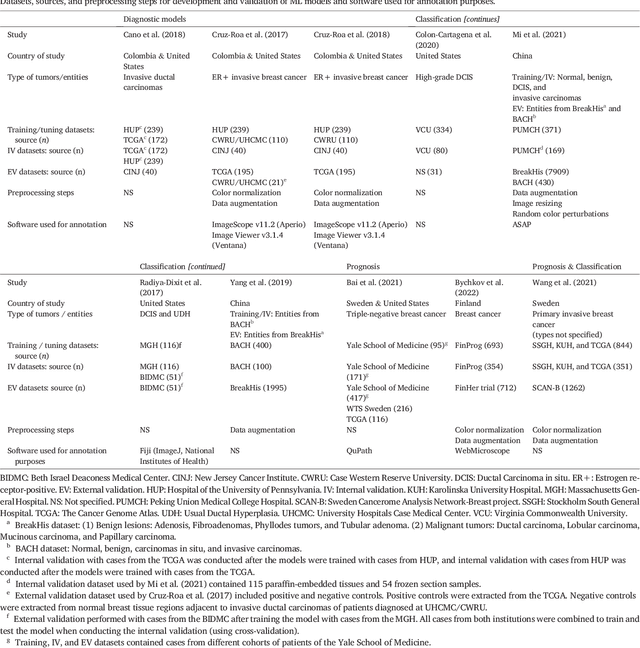
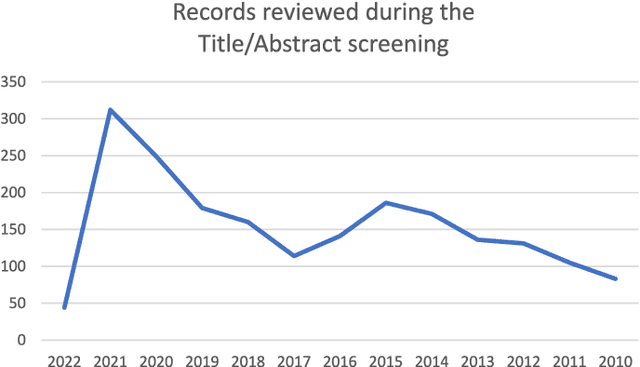
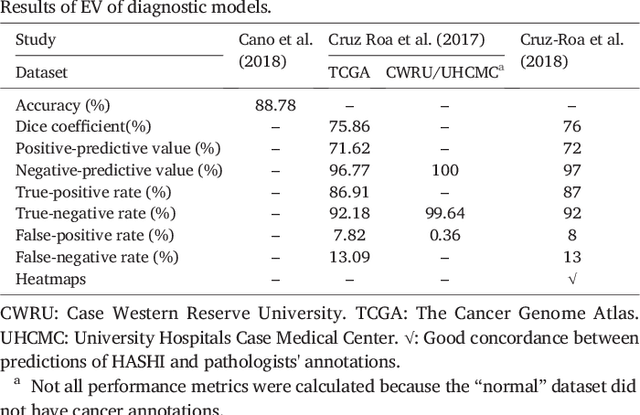
Abstract:Numerous machine learning (ML) models have been developed for breast cancer using various types of data. Successful external validation (EV) of ML models is important evidence of their generalizability. The aim of this systematic review was to assess the performance of externally validated ML models based on histopathology images for diagnosis, classification, prognosis, or treatment outcome prediction in female breast cancer. A systematic search of MEDLINE, EMBASE, CINAHL, IEEE, MICCAI, and SPIE conferences was performed for studies published between January 2010 and February 2022. The Prediction Model Risk of Bias Assessment Tool (PROBAST) was employed, and the results were narratively described. Of the 2011 non-duplicated citations, 8 journal articles and 2 conference proceedings met inclusion criteria. Three studies externally validated ML models for diagnosis, 4 for classification, 2 for prognosis, and 1 for both classification and prognosis. Most studies used Convolutional Neural Networks and one used logistic regression algorithms. For diagnostic/classification models, the most common performance metrics reported in the EV were accuracy and area under the curve, which were greater than 87% and 90%, respectively, using pathologists' annotations as ground truth. The hazard ratios in the EV of prognostic ML models were between 1.7 (95% CI, 1.2-2.6) and 1.8 (95% CI, 1.3-2.7) to predict distant disease-free survival; 1.91 (95% CI, 1.11-3.29) for recurrence, and between 0.09 (95% CI, 0.01-0.70) and 0.65 (95% CI, 0.43-0.98) for overall survival, using clinical data as ground truth. Despite EV being an important step before the clinical application of a ML model, it hasn't been performed routinely. The large variability in the training/validation datasets, methods, performance metrics, and reported information limited the comparison of the models and the analysis of their results (...)
Seeing the random forest through the decision trees. Supporting learning health systems from histopathology with machine learning models: Challenges and opportunities
Dec 06, 2023Abstract:This paper discusses some overlooked challenges faced when working with machine learning models for histopathology and presents a novel opportunity to support "Learning Health Systems" with them. Initially, the authors elaborate on these challenges after separating them according to their mitigation strategies: those that need innovative approaches, time, or future technological capabilities and those that require a conceptual reappraisal from a critical perspective. Then, a novel opportunity to support "Learning Health Systems" by integrating hidden information extracted by ML models from digitalized histopathology slides with other healthcare big data is presented.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge