Andrés Bell-Navas
CardioMOD-Net: A Modal Decomposition-Neural Network Framework for Diagnosis and Prognosis of HFpEF from Echocardiography Cine Loops
Jan 03, 2026Abstract:Introduction: Heart failure with preserved ejection fraction (HFpEF) arises from diverse comorbidities and progresses through prolonged subclinical stages, making early diagnosis and prognosis difficult. Current echocardiography-based Artificial Intelligence (AI) models focus primarily on binary HFpEF detection in humans and do not provide comorbidity-specific phenotyping or temporal estimates of disease progression towards decompensation. We aimed to develop a unified AI framework, CardioMOD-Net, to perform multiclass diagnosis and continuous prediction of HFpEF onset directly from standard echocardiography cine loops in preclinical models. Methods: Mouse echocardiography videos from four groups were used: control (CTL), hyperglycaemic (HG), obesity (OB), and systemic arterial hypertension (SAH). Two-dimensional parasternal long-axis cine loops were decomposed using Higher Order Dynamic Mode Decomposition (HODMD) to extract temporal features for downstream analysis. A shared latent representation supported Vision Transformers, one for a classifier for diagnosis and another for a regression module for predicting the age at HFpEF onset. Results: Overall diagnostic accuracy across the four groups was 65%, with all classes exceeding 50% accuracy. Misclassifications primarily reflected early-stage overlap between OB or SAH and CTL. The prognostic module achieved a root-mean-square error of 21.72 weeks for time-to-HFpEF prediction, with OB and SAH showing the most accurate estimates. Predicted HFpEF onset closely matched true distributions in all groups. Discussion: This unified framework demonstrates that multiclass phenotyping and continuous HFpEF onset prediction can be obtained from a single cine loop, even under small-data conditions. The approach offers a foundation for integrating diagnostic and prognostic modelling in preclinical HFpEF research.
A Critical Assessment of Pattern Comparisons Between POD and Autoencoders in Intraventricular Flows
Dec 22, 2025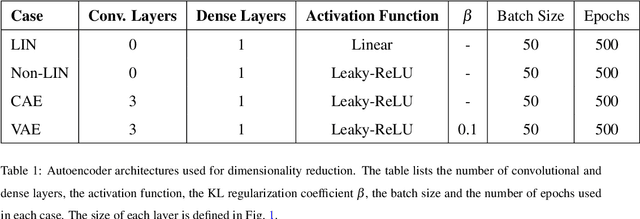
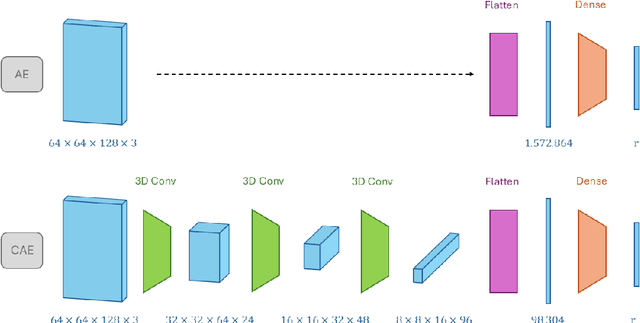
Abstract:Understanding intraventricular hemodynamics requires compact and physically interpretable representations of the underlying flow structures, as characteristic flow patterns are closely associated with cardiovascular conditions and can support early detection of cardiac deterioration. Conventional visualization of velocity or pressure fields, however, provides limited insight into the coherent mechanisms driving these dynamics. Reduced-order modeling techniques, like Proper Orthogonal Decomposition (POD) and Autoencoder (AE) architectures, offer powerful alternatives to extract dominant flow features from complex datasets. This study systematically compares POD with several AE variants (Linear, Nonlinear, Convolutional, and Variational) using left ventricular flow fields obtained from computational fluid dynamics simulations. We show that, for a suitably chosen latent dimension, AEs produce modes that become nearly orthogonal and qualitatively resemble POD modes that capture a given percentage of kinetic energy. As the number of latent modes increases, AE modes progressively lose orthogonality, leading to linear dependence, spatial redundancy, and the appearance of repeated modes with substantial high-frequency content. This degradation reduces interpretability and introduces noise-like components into AE-based reduced-order models, potentially complicating their integration with physics-based formulations or neural-network surrogates. The extent of interpretability loss varies across the AEs, with nonlinear, convolutional, and variational models exhibiting distinct behaviors in orthogonality preservation and feature localization. Overall, the results indicate that AEs can reproduce POD-like coherent structures under specific latent-space configurations, while highlighting the need for careful mode selection to ensure physically meaningful representations of cardiac flow dynamics.
Heart Failure Prediction using Modal Decomposition and Masked Autoencoders for Scarce Echocardiography Databases
Apr 10, 2025Abstract:Heart diseases constitute the main cause of international human defunction. According to the World Health Organization (WHO), approximately 18 million deaths happen each year due to precisely heart diseases. In particular, heart failures (HF) press the healthcare industry to develop systems for their early, rapid and effective prediction. In this work, an automatic system which analyses in real-time echocardiography video sequences is proposed for the challenging and more specific task of prediction of heart failure times. This system is based on a novel deep learning framework, and works in two stages. The first one transforms the data included in a database of echocardiography video sequences into a machine learning-compatible collection of annotated images which can be used in the training phase of any kind of machine learning-based framework, including a deep learning one. This initial stage includes the use of the Higher Order Dynamic Mode Decomposition (HODMD) algorithm for both data augmentation and feature extraction. The second stage is focused on building and training a Vision Transformer (ViT). Self-supervised learning (SSL) methods, which have been so far barely explored in the literature about heart failure prediction, are applied to effectively train the ViT from scratch, even with scarce databases of echocardiograms. The designed neural network analyses images from echocardiography sequences to estimate the time in which a heart failure will happen. The results obtained show the efficacy of the HODMD algorithm and the superiority of the proposed system with respect to several established ViT and Convolutional Neural Network (CNN) architectures.
Automatic Cardiac Pathology Recognition in Echocardiography Images Using Higher Order Dynamic Mode Decomposition and a Vision Transformer for Small Datasets
Apr 30, 2024Abstract:Heart diseases are the main international cause of human defunction. According to the WHO, nearly 18 million people decease each year because of heart diseases. Also considering the increase of medical data, much pressure is put on the health industry to develop systems for early and accurate heart disease recognition. In this work, an automatic cardiac pathology recognition system based on a novel deep learning framework is proposed, which analyses in real-time echocardiography video sequences. The system works in two stages. The first one transforms the data included in a database of echocardiography sequences into a machine-learning-compatible collection of annotated images which can be used in the training stage of any kind of machine learning-based framework, and more specifically with deep learning. This includes the use of the Higher Order Dynamic Mode Decomposition (HODMD) algorithm, for the first time to the authors' knowledge, for both data augmentation and feature extraction in the medical field. The second stage is focused on building and training a Vision Transformer (ViT), barely explored in the related literature. The ViT is adapted for an effective training from scratch, even with small datasets. The designed neural network analyses images from an echocardiography sequence to predict the heart state. The results obtained show the superiority of the proposed system and the efficacy of the HODMD algorithm, even outperforming pretrained Convolutional Neural Networks (CNNs), which are so far the method of choice in the literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge