Nourelhouda Groun
EigenHearts: Cardiac Diseases Classification Using EigenFaces Approach
Nov 25, 2024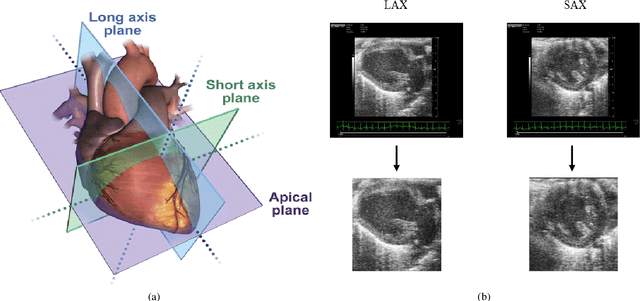
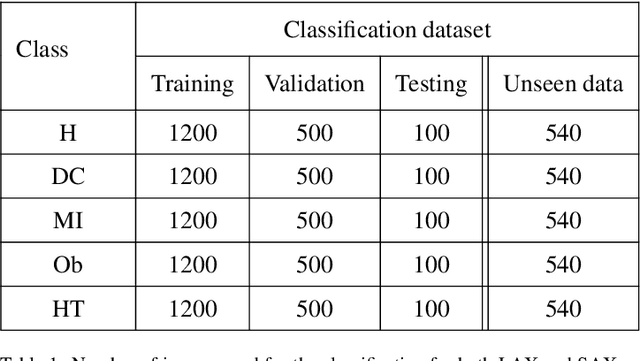
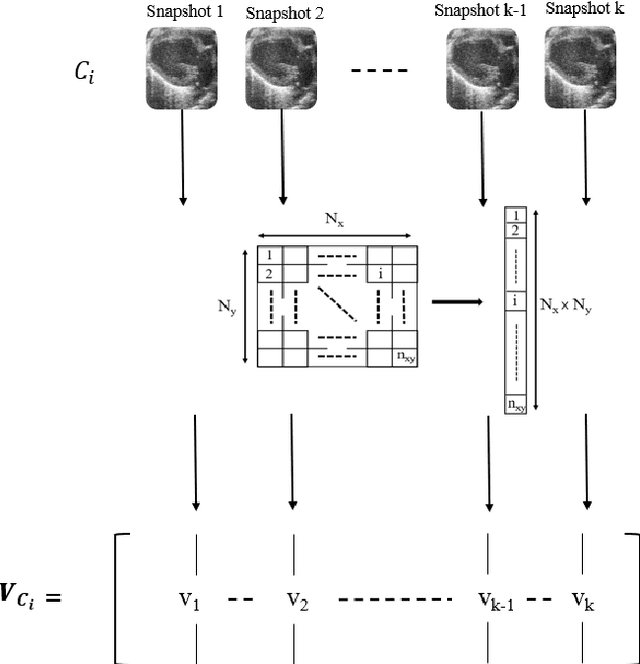
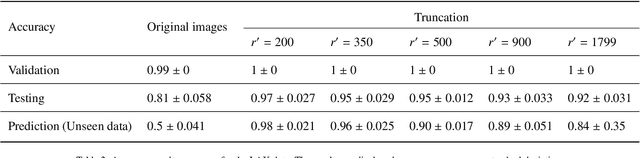
Abstract:In the realm of cardiovascular medicine, medical imaging plays a crucial role in accurately classifying cardiac diseases and making precise diagnoses. However, the field faces significant challenges when integrating data science techniques, as a significant volume of images is required for these techniques. As a consequence, it is necessary to investigate different avenues to overcome this challenge. In this contribution, we offer an innovative tool to conquer this limitation. In particular, we delve into the application of a well recognized method known as the EigenFaces approach to classify cardiac diseases. This approach was originally motivated for efficiently representing pictures of faces using principal component analysis, which provides a set of eigenvectors (aka eigenfaces), explaining the variation between face images. As this approach proven to be efficient for face recognition, it motivated us to explore its efficiency on more complicated data bases. In particular, we integrate this approach, with convolutional neural networks (CNNs) to classify echocardiography images taken from mice in five distinct cardiac conditions (healthy, diabetic cardiomyopathy, myocardial infarction, obesity and TAC hypertension). Performing a preprocessing step inspired from the eigenfaces approach on the echocardiography datasets, yields sets of pod modes, which we will call eigenhearts. To demonstrate the proposed approach, we compare two testcases: (i) supplying the CNN with the original images directly, (ii) supplying the CNN with images projected into the obtained pod modes. The results show a substantial and noteworthy enhancement when employing SVD for pre-processing, with classification accuracy increasing by approximately 50%.
A Novel Data Augmentation Tool for Enhancing Machine Learning Classification: A New Application of the Higher Order Dynamic Mode Decomposition for Improved Cardiac Disease Identification
Nov 24, 2024



Abstract:In this work, a data-driven, modal decomposition method, the higher order dynamic mode decomposition (HODMD), is combined with a convolutional neural network (CNN) in order to improve the classification accuracy of several cardiac diseases using echocardiography images. The HODMD algorithm is used first as feature extraction technique for the echocardiography datasets, taken from both healthy mice and mice afflicted by different cardiac diseases (Diabetic Cardiomyopathy, Obesity, TAC Hypertrophy and Myocardial Infarction). A total number of 130 echocardiography datasets are used in this work. The dominant features related to each cardiac disease were identified and represented by the HODMD algorithm as a set of DMD modes, which then are used as the input to the CNN. In a way, the database dimension was augmented, hence HODMD has been used, for the first time to the authors knowledge, for data augmentation in the machine learning framework. Six sets of the original echocardiography databases were hold out to be used as unseen data to test the performance of the CNN. In order to demonstrate the efficiency of the HODMD technique, two testcases are studied: the CNN is first trained using the original echocardiography images only, and second training the CNN using a combination of the original images and the DMD modes. The classification performance of the designed trained CNN shows that combining the original images with the DMD modes improves the results in all the testcases, as it improves the accuracy by up to 22%. These results show the great potential of using the HODMD algorithm as a data augmentation technique.
Automatic Cardiac Pathology Recognition in Echocardiography Images Using Higher Order Dynamic Mode Decomposition and a Vision Transformer for Small Datasets
Apr 30, 2024Abstract:Heart diseases are the main international cause of human defunction. According to the WHO, nearly 18 million people decease each year because of heart diseases. Also considering the increase of medical data, much pressure is put on the health industry to develop systems for early and accurate heart disease recognition. In this work, an automatic cardiac pathology recognition system based on a novel deep learning framework is proposed, which analyses in real-time echocardiography video sequences. The system works in two stages. The first one transforms the data included in a database of echocardiography sequences into a machine-learning-compatible collection of annotated images which can be used in the training stage of any kind of machine learning-based framework, and more specifically with deep learning. This includes the use of the Higher Order Dynamic Mode Decomposition (HODMD) algorithm, for the first time to the authors' knowledge, for both data augmentation and feature extraction in the medical field. The second stage is focused on building and training a Vision Transformer (ViT), barely explored in the related literature. The ViT is adapted for an effective training from scratch, even with small datasets. The designed neural network analyses images from an echocardiography sequence to predict the heart state. The results obtained show the superiority of the proposed system and the efficacy of the HODMD algorithm, even outperforming pretrained Convolutional Neural Networks (CNNs), which are so far the method of choice in the literature.
A Novel Data-Driven Method for the Analysis and Reconstruction of Cardiac Cine MRI
May 24, 2022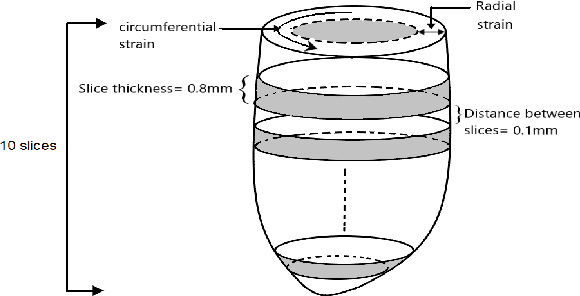
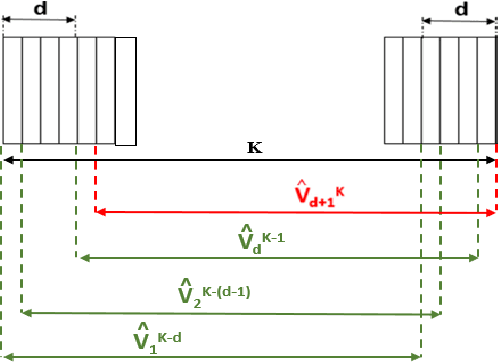
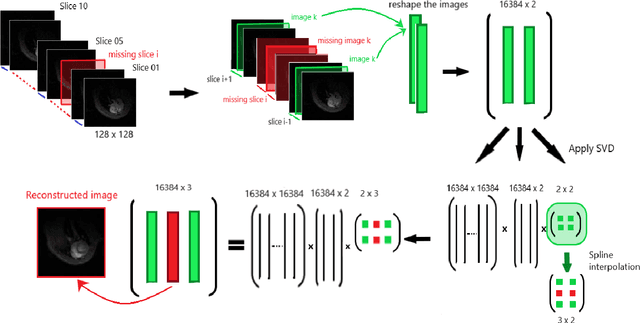
Abstract:Cardiac cine magnetic resonance imaging (MRI) can be considered the optimal criterion for measuring cardiac function. This imaging technique can provide us with detailed information about cardiac structure, tissue composition and even blood flow. This work considers the application of the higher order dynamic mode decomposition (HODMD) method to a set of MR images of a heart, with the ultimate goal of identifying the main patterns and frequencies driving the heart dynamics. A novel algorithm based on singular value decomposition combined with HODMD is introduced, providing a three-dimensional reconstruction of the heart. This algorithm is applied (i) to reconstruct corrupted or missing images, and (ii) to build a reduced order model of the heart dynamics.
Higher Order Dynamic Mode Decomposition: from Fluid Dynamics to Heart Disease Analysis
Jan 09, 2022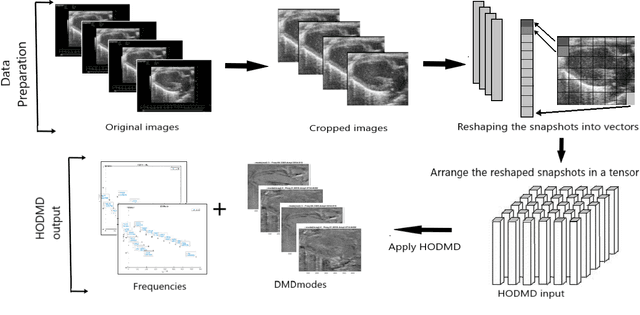
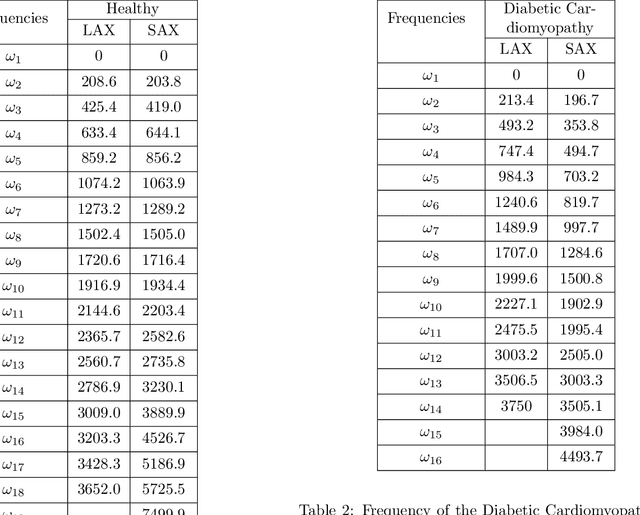
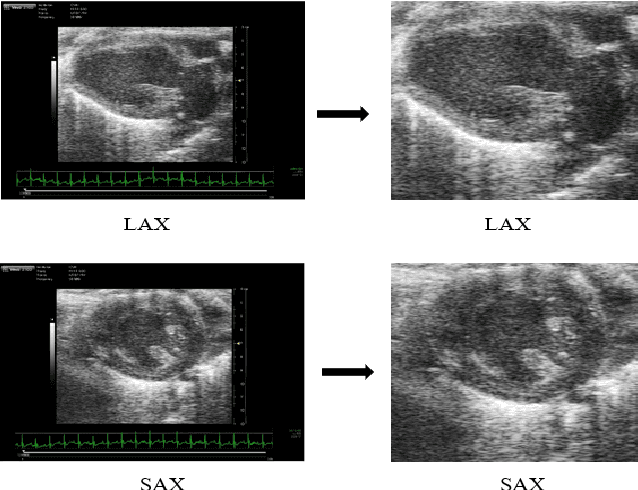
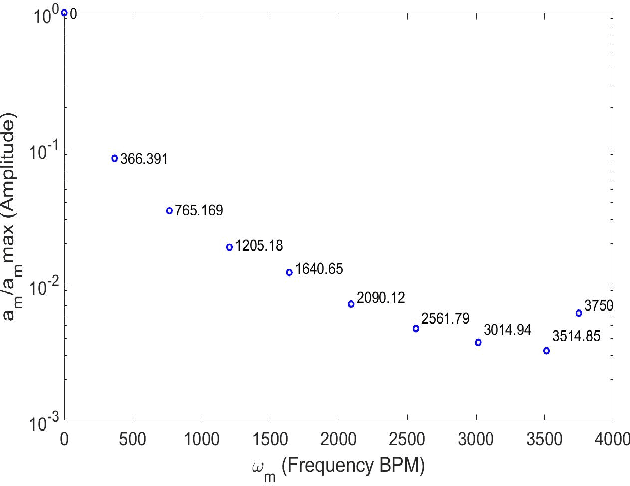
Abstract:In this work, we study in detail the performance of Higher Order Dynamic Mode Decomposition (HODMD) technique when applied to echocardiography images. HODMD is a data-driven method generally used in fluid dynamics and in the analysis of complex non-linear dynamical systems modeling several complex industrial applications. In this paper we apply HODMD, for the first time to the authors knowledge, for patterns recognition in echocardiography, specifically, echocardiography data taken from several mice, either in healthy conditions or afflicted by different cardiac diseases. We exploit the HODMD advantageous properties in dynamics identification and noise cleaning to identify the relevant frequencies and coherent patterns for each one of the diseases. The echocardiography datasets consist of video loops taken with respect to a long axis view (LAX) and a short axis view (SAX), where each video loop covers at least three cardiac cycles, formed by (at most) 300 frames each (called snapshots). The proposed algorithm, using only a maximum quantity of 200 snapshots, was able to capture two branches of frequencies, representing the heart rate and respiratory rate. Additionally, the algorithm provided a number of modes, which represent the dominant features and patterns in the different echocardiography images, also related to the heart and the lung. Six datasets were analyzed: one echocardiography taken from a healthy subject and five different sets of echocardiography taken from subjects with either Diabetic Cardiomyopathy, Obesity, SFSR4 Hypertrophy, TAC Hypertrophy or Myocardial Infarction. The results show that HODMD is robust and a suitable tool to identify characteristic patterns able to classify the different pathologies studied.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge