Alireza Akhondi-Asl
Bayesian Dynamic DAG Learning: Application in Discovering Dynamic Effective Connectome of Brain
Sep 07, 2023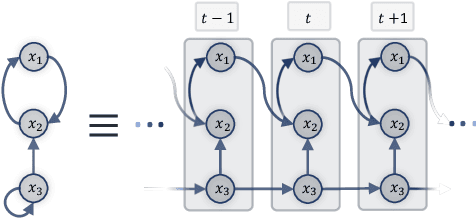

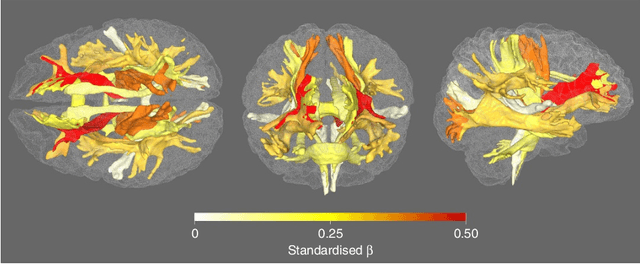
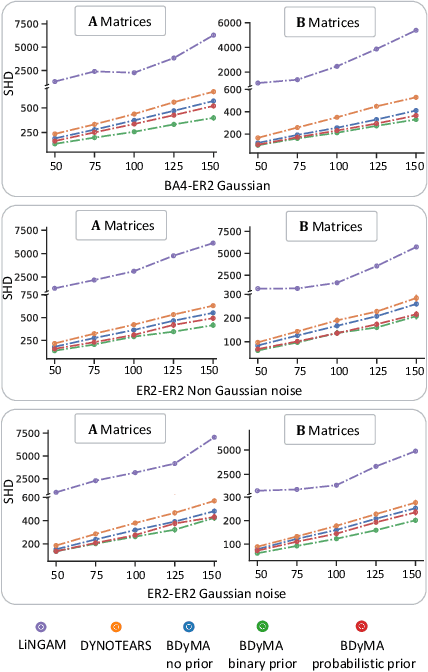
Abstract:Understanding the complex mechanisms of the brain can be unraveled by extracting the Dynamic Effective Connectome (DEC). Recently, score-based Directed Acyclic Graph (DAG) discovery methods have shown significant improvements in extracting the causal structure and inferring effective connectivity. However, learning DEC through these methods still faces two main challenges: one with the fundamental impotence of high-dimensional dynamic DAG discovery methods and the other with the low quality of fMRI data. In this paper, we introduce Bayesian Dynamic DAG learning with M-matrices Acyclicity characterization \textbf{(BDyMA)} method to address the challenges in discovering DEC. The presented dynamic causal model enables us to discover bidirected edges as well. Leveraging an unconstrained framework in the BDyMA method leads to more accurate results in detecting high-dimensional networks, achieving sparser outcomes, making it particularly suitable for extracting DEC. Additionally, the score function of the BDyMA method allows the incorporation of prior knowledge into the process of dynamic causal discovery which further enhances the accuracy of results. Comprehensive simulations on synthetic data and experiments on Human Connectome Project (HCP) data demonstrate that our method can handle both of the two main challenges, yielding more accurate and reliable DEC compared to state-of-the-art and baseline methods. Additionally, we investigate the trustworthiness of DTI data as prior knowledge for DEC discovery and show the improvements in DEC discovery when the DTI data is incorporated into the process.
Brain Effective Connectome based on fMRI and DTI Data: Bayesian Causal Learning and Assessment
Feb 21, 2023



Abstract:Neuroscientific studies aim to find an accurate and reliable brain Effective Connectome (EC). Although current EC discovery methods have contributed to our understanding of brain organization, their performances are severely constrained by the short sample size and poor temporal resolution of fMRI data, and high dimensionality of the brain connectome. By leveraging the DTI data as prior knowledge, we introduce two Bayesian causal discovery frameworks -- the Bayesian GOLEM (BGOLEM) and Bayesian FGES (BFGES) methods -- that offer significantly more accurate and reliable ECs and address the shortcomings of the existing causal discovery methods in discovering ECs based on only fMRI data. Through a series of simulation studies on synthetic and hybrid (DTI of the Human Connectome Project (HCP) subjects and synthetic fMRI) data, we demonstrate the effectiveness of the proposed methods in discovering EC. To numerically assess the improvement in the accuracy of ECs with our method on empirical data, we first introduce the Pseudo False Discovery Rate (PFDR) as a new computational accuracy metric for causal discovery in the brain. We show that our Bayesian methods achieve higher accuracy than traditional methods on HCP data. Additionally, we measure the reliability of discovered ECs using the Rogers-Tanimoto index for test-retest data and show that our Bayesian methods provide significantly more reproducible ECs than traditional methods. Overall, our study's numerical and graphical results highlight the potential for these frameworks to advance our understanding of brain function and organization significantly.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge