Alice Schoenauer-Sebag
Multi-Domain Adversarial Learning
Mar 21, 2019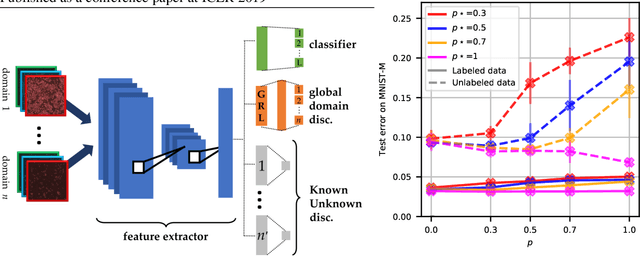



Abstract:Multi-domain learning (MDL) aims at obtaining a model with minimal average risk across multiple domains. Our empirical motivation is automated microscopy data, where cultured cells are imaged after being exposed to known and unknown chemical perturbations, and each dataset displays significant experimental bias. This paper presents a multi-domain adversarial learning approach, MuLANN, to leverage multiple datasets with overlapping but distinct class sets, in a semi-supervised setting. Our contributions include: i) a bound on the average- and worst-domain risk in MDL, obtained using the H-divergence; ii) a new loss to accommodate semi-supervised multi-domain learning and domain adaptation; iii) the experimental validation of the approach, improving on the state of the art on two standard image benchmarks, and a novel bioimage dataset, Cell.
* Accepted at ICLR'19
Stochastic Gradient Descent: Going As Fast As Possible But Not Faster
Sep 05, 2017Abstract:When applied to training deep neural networks, stochastic gradient descent (SGD) often incurs steady progression phases, interrupted by catastrophic episodes in which loss and gradient norm explode. A possible mitigation of such events is to slow down the learning process. This paper presents a novel approach to control the SGD learning rate, that uses two statistical tests. The first one, aimed at fast learning, compares the momentum of the normalized gradient vectors to that of random unit vectors and accordingly gracefully increases or decreases the learning rate. The second one is a change point detection test, aimed at the detection of catastrophic learning episodes; upon its triggering the learning rate is instantly halved. Both abilities of speeding up and slowing down the learning rate allows the proposed approach, called SALeRA, to learn as fast as possible but not faster. Experiments on standard benchmarks show that SALeRA performs well in practice, and compares favorably to the state of the art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge