Alexander Hann
In-depth Assessment of an Interactive Graph-based Approach for the Segmentation for Pancreatic Metastasis in Ultrasound Acquisitions of the Liver with two Specialists in Internal Medicine
Mar 12, 2018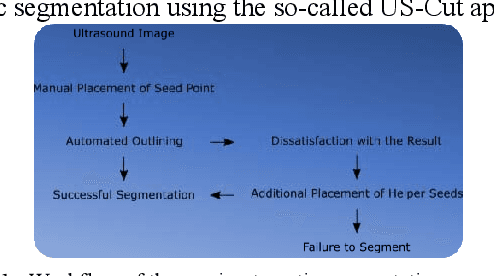
Abstract:The manual outlining of hepatic metastasis in (US) ultrasound acquisitions from patients suffering from pancreatic cancer is common practice. However, such pure manual measurements are often very time consuming, and the results repeatedly differ between the raters. In this contribution, we study the in-depth assessment of an interactive graph-based approach for the segmentation for pancreatic metastasis in US images of the liver with two specialists in Internal Medicine. Thereby, evaluating the approach with over one hundred different acquisitions of metastases. The two physicians or the algorithm had never assessed the acquisitions before the evaluation. In summary, the physicians first performed a pure manual outlining followed by an algorithmic segmentation over one month later. As a result, the experts satisfied in up to ninety percent of algorithmic segmentation results. Furthermore, the algorithmic segmentation was much faster than manual outlining and achieved a median Dice Similarity Coefficient (DSC) of over eighty percent. Ultimately, the algorithm enables a fast and accurate segmentation of liver metastasis in clinical US images, which can support the manual outlining in daily practice.
Algorithm guided outlining of 105 pancreatic cancer liver metastases in Ultrasound
Oct 09, 2017



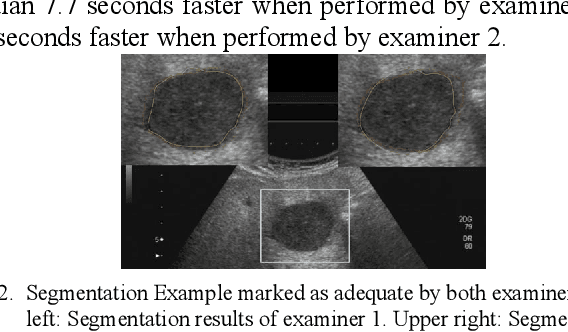
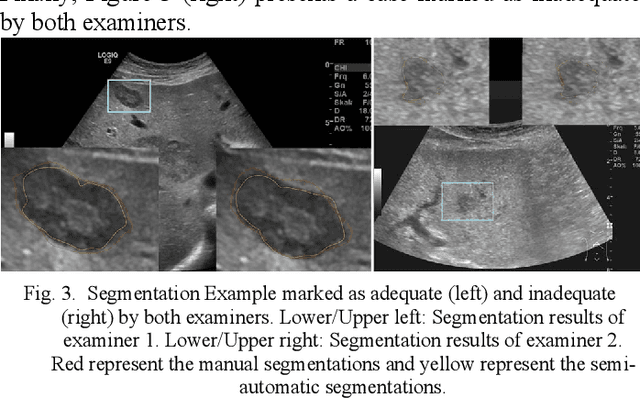
Abstract:Manual segmentation of hepatic metastases in ultrasound images acquired from patients suffering from pancreatic cancer is common practice. Semiautomatic measurements promising assistance in this process are often assessed using a small number of lesions performed by examiners who already know the algorithm. In this work, we present the application of an algorithm for the segmentation of liver metastases due to pancreatic cancer using a set of 105 different images of metastases. The algorithm and the two examiners had never assessed the images before. The examiners first performed a manual segmentation and, after five weeks, a semiautomatic segmentation using the algorithm. They were satisfied in up to 90% of the cases with the semiautomatic segmentation results. Using the algorithm was significantly faster and resulted in a median Dice similarity score of over 80%. Estimation of the inter-operator variability by using the intra class correlation coefficient was good with 0.8. In conclusion, the algorithm facilitates fast and accurate segmentation of liver metastases, comparable to the current gold standard of manual segmentation.
* 7 pages, 3 Figures, 3 Tables, 46 References
Interactive Outlining of Pancreatic Cancer Liver Metastases in Ultrasound Images
Apr 18, 2017



Abstract:Ultrasound (US) is the most commonly used liver imaging modality worldwide. Due to its low cost, it is increasingly used in the follow-up of cancer patients with metastases localized in the liver. In this contribution, we present the results of an interactive segmentation approach for liver metastases in US acquisitions. A (semi-) automatic segmentation is still very challenging because of the low image quality and the low contrast between the metastasis and the surrounding liver tissue. Thus, the state of the art in clinical practice is still manual measurement and outlining of the metastases in the US images. We tackle the problem by providing an interactive segmentation approach providing real-time feedback of the segmentation results. The approach has been evaluated with typical US acquisitions from the clinical routine, and the datasets consisted of pancreatic cancer metastases. Even for difficult cases, satisfying segmentations results could be achieved because of the interactive real-time behavior of the approach. In total, 40 clinical images have been evaluated with our method by comparing the results against manual ground truth segmentations. This evaluation yielded to an average Dice Score of 85% and an average Hausdorff Distance of 13 pixels.
* 15 pages, 16 figures, 2 tables, 58 references
US-Cut: Interactive Algorithm for rapid Detection and Segmentation of Liver Tumors in Ultrasound Acquisitions
Mar 02, 2016Abstract:Ultrasound (US) is the most commonly used liver imaging modality worldwide. It plays an important role in follow-up of cancer patients with liver metastases. We present an interactive segmentation approach for liver tumors in US acquisitions. Due to the low image quality and the low contrast between the tumors and the surrounding tissue in US images, the segmentation is very challenging. Thus, the clinical practice still relies on manual measurement and outlining of the tumors in the US images. We target this problem by applying an interactive segmentation algorithm to the US data, allowing the user to get real-time feedback of the segmentation results. The algorithm has been developed and tested hand-in-hand by physicians and computer scientists to make sure a future practical usage in a clinical setting is feasible. To cover typical acquisitions from the clinical routine, the approach has been evaluated with dozens of datasets where the tumors are hyperechoic (brighter), hypoechoic (darker) or isoechoic (similar) in comparison to the surrounding liver tissue. Due to the interactive real-time behavior of the approach, it was possible even in difficult cases to find satisfying segmentations of the tumors within seconds and without parameter settings, and the average tumor deviation was only 1.4mm compared with manual measurements. However, the long term goal is to ease the volumetric acquisition of liver tumors in order to evaluate for treatment response. Additional aim is the registration of intraoperative US images via the interactive segmentations to the patient's pre-interventional CT acquisitions.
* 6 pages, 6 figures, 1 table, 32 references
Interactive Volumetry Of Liver Ablation Zones
Oct 21, 2015



Abstract:Percutaneous radiofrequency ablation (RFA) is a minimally invasive technique that destroys cancer cells by heat. The heat results from focusing energy in the radiofrequency spectrum through a needle. Amongst others, this can enable the treatment of patients who are not eligible for an open surgery. However, the possibility of recurrent liver cancer due to incomplete ablation of the tumor makes post-interventional monitoring via regular follow-up scans mandatory. These scans have to be carefully inspected for any conspicuousness. Within this study, the RF ablation zones from twelve post-interventional CT acquisitions have been segmented semi-automatically to support the visual inspection. An interactive, graph-based contouring approach, which prefers spherically shaped regions, has been applied. For the quantitative and qualitative analysis of the algorithm's results, manual slice-by-slice segmentations produced by clinical experts have been used as the gold standard (which have also been compared among each other). As evaluation metric for the statistical validation, the Dice Similarity Coefficient (DSC) has been calculated. The results show that the proposed tool provides lesion segmentation with sufficient accuracy much faster than manual segmentation. The visual feedback and interactivity make the proposed tool well suitable for the clinical workflow.
* 18 pages, 15 figures, 8 tables, 57 references
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge