Ahmad Hussein
Vision Transformers with Autoencoders and Explainable AI for Cancer Patient Risk Stratification Using Whole Slide Imaging
Apr 08, 2025



Abstract:Cancer remains one of the leading causes of mortality worldwide, necessitating accurate diagnosis and prognosis. Whole Slide Imaging (WSI) has become an integral part of clinical workflows with advancements in digital pathology. While various studies have utilized WSIs, their extracted features may not fully capture the most relevant pathological information, and their lack of interpretability limits clinical adoption. In this paper, we propose PATH-X, a framework that integrates Vision Transformers (ViT) and Autoencoders with SHAP (Shapley Additive Explanations) to enhance model explainability for patient stratification and risk prediction using WSIs from The Cancer Genome Atlas (TCGA). A representative image slice is selected from each WSI, and numerical feature embeddings are extracted using Google's pre-trained ViT. These features are then compressed via an autoencoder and used for unsupervised clustering and classification tasks. Kaplan-Meier survival analysis is applied to evaluate stratification into two and three risk groups. SHAP is used to identify key contributing features, which are mapped onto histopathological slices to provide spatial context. PATH-X demonstrates strong performance in breast and glioma cancers, where a sufficient number of WSIs enabled robust stratification. However, performance in lung cancer was limited due to data availability, emphasizing the need for larger datasets to enhance model reliability and clinical applicability.
Explainable AI Methods for Multi-Omics Analysis: A Survey
Oct 15, 2024

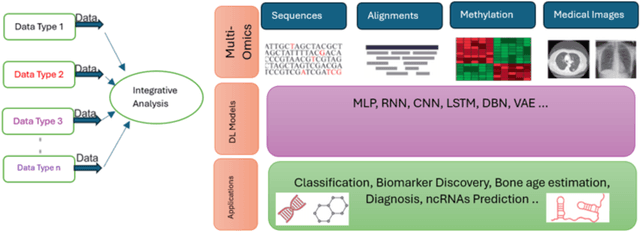
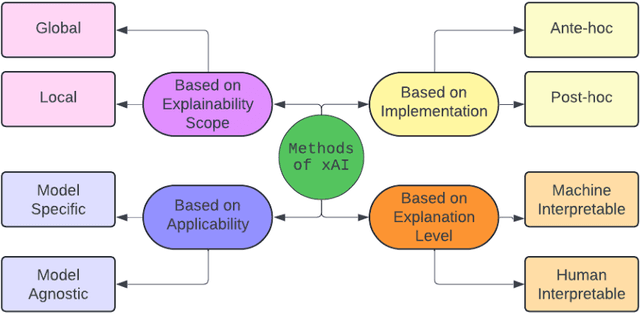
Abstract:Advancements in high-throughput technologies have led to a shift from traditional hypothesis-driven methodologies to data-driven approaches. Multi-omics refers to the integrative analysis of data derived from multiple 'omes', such as genomics, proteomics, transcriptomics, metabolomics, and microbiomics. This approach enables a comprehensive understanding of biological systems by capturing different layers of biological information. Deep learning methods are increasingly utilized to integrate multi-omics data, offering insights into molecular interactions and enhancing research into complex diseases. However, these models, with their numerous interconnected layers and nonlinear relationships, often function as black boxes, lacking transparency in decision-making processes. To overcome this challenge, explainable artificial intelligence (xAI) methods are crucial for creating transparent models that allow clinicians to interpret and work with complex data more effectively. This review explores how xAI can improve the interpretability of deep learning models in multi-omics research, highlighting its potential to provide clinicians with clear insights, thereby facilitating the effective application of such models in clinical settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge