Abigail Hickok
Discrete scalar curvature as a weighted sum of Ollivier-Ricci curvatures
Oct 06, 2025

Abstract:We study the relationship between discrete analogues of Ricci and scalar curvature that are defined for point clouds and graphs. In the discrete setting, Ricci curvature is replaced by Ollivier-Ricci curvature. Scalar curvature can be computed as the trace of Ricci curvature for a Riemannian manifold; this motivates a new definition of a scalar version of Ollivier-Ricci curvature. We show that our definition converges to scalar curvature for nearest neighbor graphs obtained by sampling from a manifold. We also prove some new results about the convergence of Ollivier-Ricci curvature to Ricci curvature.
EmbedOR: Provable Cluster-Preserving Visualizations with Curvature-Based Stochastic Neighbor Embeddings
Sep 03, 2025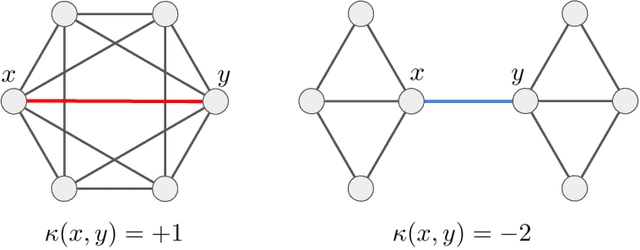
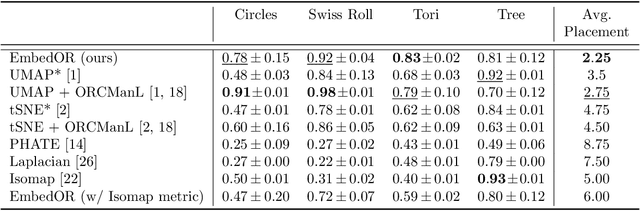
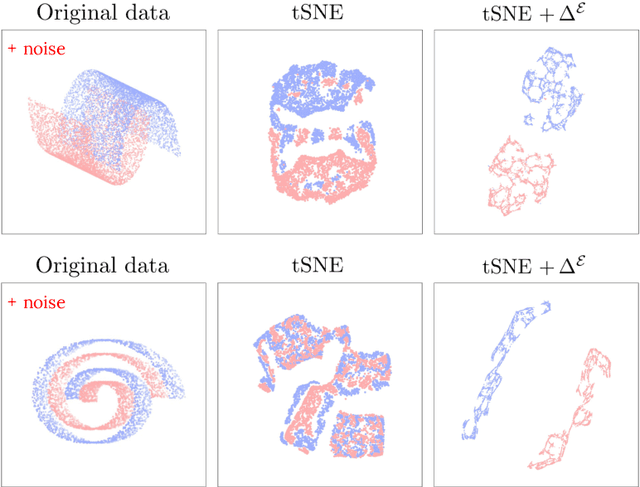

Abstract:Stochastic Neighbor Embedding (SNE) algorithms like UMAP and tSNE often produce visualizations that do not preserve the geometry of noisy and high dimensional data. In particular, they can spuriously separate connected components of the underlying data submanifold and can fail to find clusters in well-clusterable data. To address these limitations, we propose EmbedOR, a SNE algorithm that incorporates discrete graph curvature. Our algorithm stochastically embeds the data using a curvature-enhanced distance metric that emphasizes underlying cluster structure. Critically, we prove that the EmbedOR distance metric extends consistency results for tSNE to a much broader class of datasets. We also describe extensive experiments on synthetic and real data that demonstrate the visualization and geometry-preservation capabilities of EmbedOR. We find that, unlike other SNE algorithms and UMAP, EmbedOR is much less likely to fragment continuous, high-density regions of the data. Finally, we demonstrate that the EmbedOR distance metric can be used as a tool to annotate existing visualizations to identify fragmentation and provide deeper insight into the underlying geometry of the data.
Recovering Manifold Structure Using Ollivier-Ricci Curvature
Oct 02, 2024



Abstract:We introduce ORC-ManL, a new algorithm to prune spurious edges from nearest neighbor graphs using a criterion based on Ollivier-Ricci curvature and estimated metric distortion. Our motivation comes from manifold learning: we show that when the data generating the nearest-neighbor graph consists of noisy samples from a low-dimensional manifold, edges that shortcut through the ambient space have more negative Ollivier-Ricci curvature than edges that lie along the data manifold. We demonstrate that our method outperforms alternative pruning methods and that it significantly improves performance on many downstream geometric data analysis tasks that use nearest neighbor graphs as input. Specifically, we evaluate on manifold learning, persistent homology, dimension estimation, and others. We also show that ORC-ManL can be used to improve clustering and manifold learning of single-cell RNA sequencing data. Finally, we provide empirical convergence experiments that support our theoretical findings.
An Intrinsic Approach to Scalar-Curvature Estimation for Point Clouds
Aug 04, 2023



Abstract:We introduce an intrinsic estimator for the scalar curvature of a data set presented as a finite metric space. Our estimator depends only on the metric structure of the data and not on an embedding in $\mathbb{R}^n$. We show that the estimator is consistent in the sense that for points sampled from a probability measure on a compact Riemannian manifold, the estimator converges to the scalar curvature as the number of points increases. To justify its use in applications, we show that the estimator is stable with respect to perturbations of the metric structure, e.g., noise in the sample or error estimating the intrinsic metric. We validate our estimator experimentally on synthetic data that is sampled from manifolds with specified curvature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge