Abdul Karim
Toxicity Prediction by Multimodal Deep Learning
Jul 19, 2019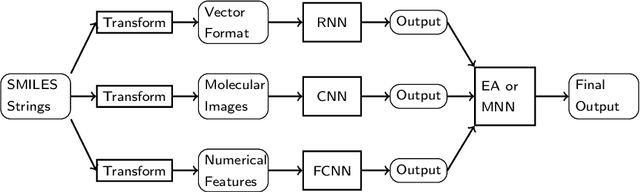
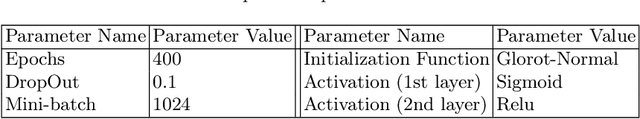
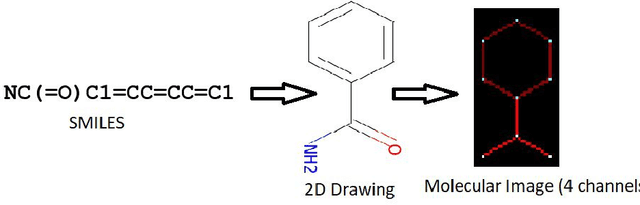
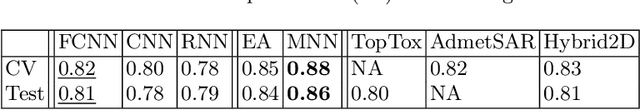
Abstract:Prediction of toxicity levels of chemical compounds is an important issue in Quantitative Structure-Activity Relationship (QSAR) modeling. Although toxicity prediction has achieved significant progress in recent times through deep learning, prediction accuracy levels obtained by even very recent methods are not yet very high. We propose a multimodal deep learning method using multiple heterogeneous neural network types and data representations. We represent chemical compounds by strings, images, and numerical features. We train fully connected, convolutional, and recurrent neural networks and their ensembles. Each data representation or neural network type has its own strengths and weaknesses. Our motivation is to obtain a collective performance that could go beyond individual performance of each data representation or each neural network type. On a standard toxicity benchmark, our proposed method obtains significantly better accuracy levels than that by the state-of-the-art toxicity prediction methods.
* Preprint Version
Efficient Toxicity Prediction via Simple Features Using Shallow Neural Networks and Decision Trees
Jan 26, 2019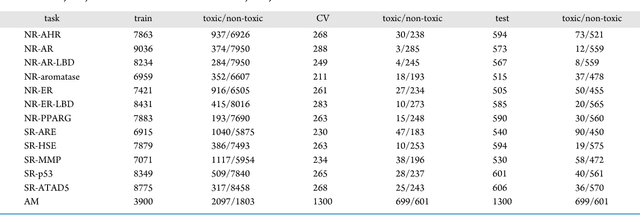
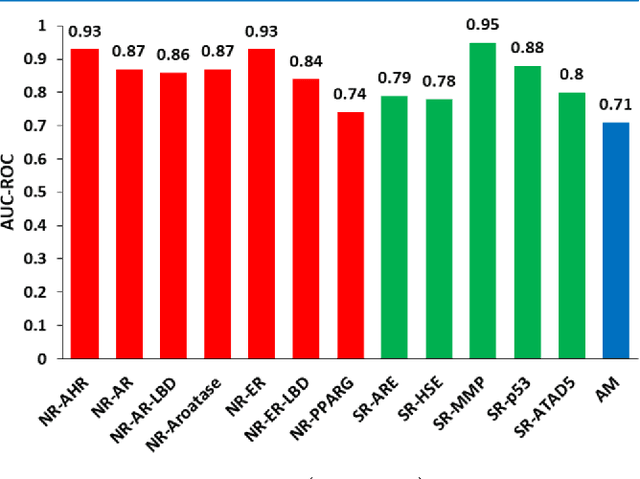
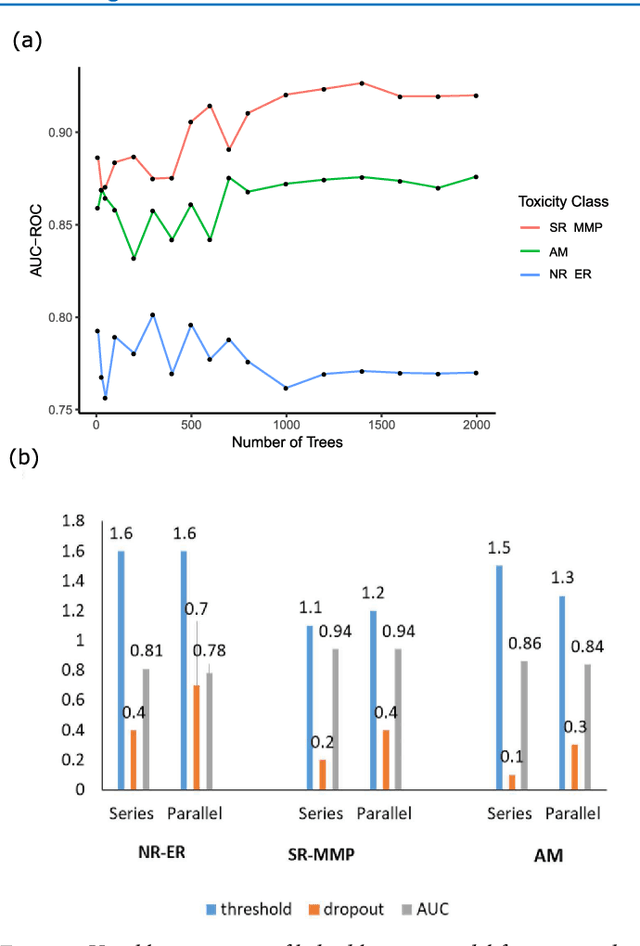
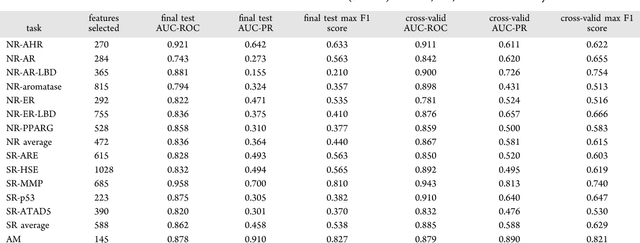
Abstract:Toxicity prediction of chemical compounds is a grand challenge. Lately, it achieved significant progress in accuracy but using a huge set of features, implementing a complex blackbox technique such as a deep neural network, and exploiting enormous computational resources. In this paper, we strongly argue for the models and methods that are simple in machine learning characteristics, efficient in computing resource usage, and powerful to achieve very high accuracy levels. To demonstrate this, we develop a single task-based chemical toxicity prediction framework using only 2D features that are less compute intensive. We effectively use a decision tree to obtain an optimum number of features from a collection of thousands of them. We use a shallow neural network and jointly optimize it with decision tree taking both network parameters and input features into account. Our model needs only a minute on a single CPU for its training while existing methods using deep neural networks need about 10 min on NVidia Tesla K40 GPU. However, we obtain similar or better performance on several toxicity benchmark tasks. We also develop a cumulative feature ranking method which enables us to identify features that can help chemists perform prescreening of toxic compounds effectively.
Machine Learning Interpretability: A Science rather than a tool
Jul 25, 2018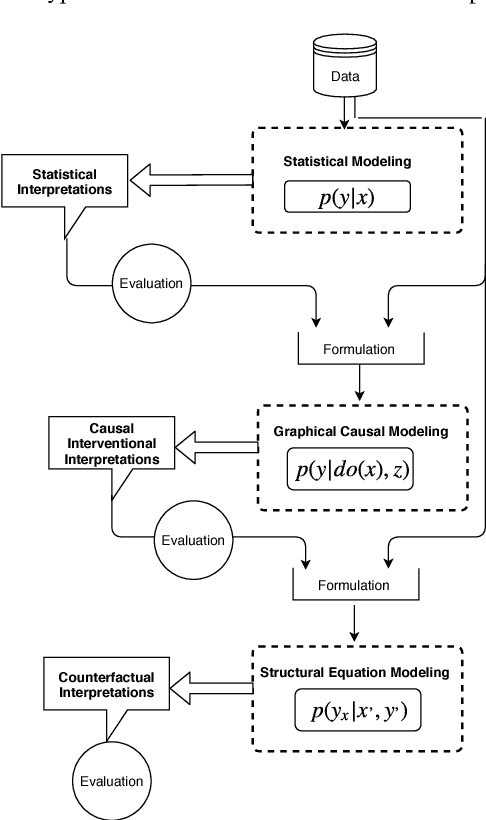
Abstract:The term "interpretability" is oftenly used by machine learning researchers each with their own intuitive understanding of it. There is no universal well agreed upon definition of interpretability in machine learning. As any type of science discipline is mainly driven by the set of formulated questions rather than by different tools in that discipline, e.g. astrophysics is the discipline that learns the composition of stars, not as the discipline that use the spectroscopes. Similarly, we propose that machine learning interpretability should be a discipline that answers specific questions related to interpretability. These questions can be of statistical, causal and counterfactual nature. Therefore, there is a need to look into the interpretability problem of machine learning in the context of questions that need to be addressed rather than different tools. We discuss about a hypothetical interpretability framework driven by a question based scientific approach rather than some specific machine learning model. Using a question based notion of interpretability, we can step towards understanding the science of machine learning rather than its engineering. This notion will also help us understanding any specific problem more in depth rather than relying solely on machine learning methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge