Aashis Khanal
Fully Automated Artery-Vein ratio and vascular tortuosity measurement in retinal fundus images
Jan 04, 2023Abstract:Accurate measurements of abnormalities like Artery-Vein ratio and tortuosity in fundus images is an actively researched task. Most of the research seems to compute such features independently. However, in this work, we have devised a fully automated technique to measure any vascular abnormalities. This paper is a follow-up paper on vessel topology estimation and extraction, we use the extracted topology to perform A-V state-of-the-art Artery-Vein classification, AV ratio calculation, and vessel tortuosity measurement, all fully automated. Existing techniques tend to only work on the partial region, but we extract the complete vascular structure. We have shown the usability of this topology by extracting two of the most important vascular features; Artery-Vein ratio, and vessel tortuosity.
Fully Automated Tree Topology Estimation and Artery-Vein Classification
Feb 04, 2022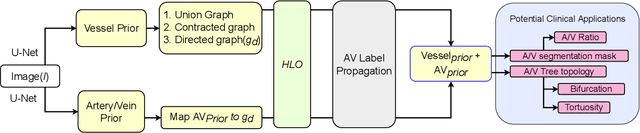
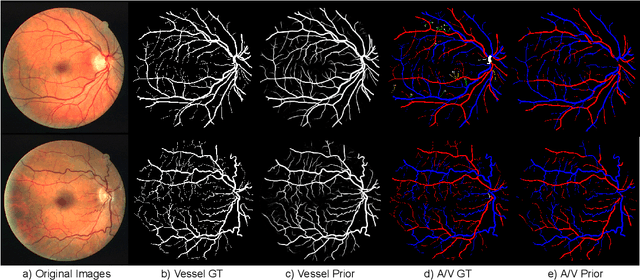
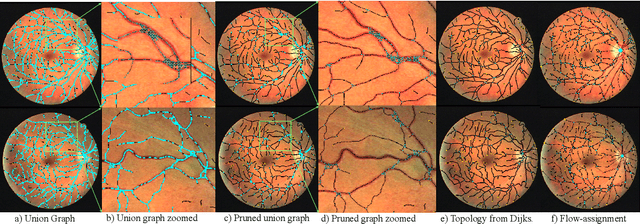
Abstract:We present a fully automatic technique for extracting the retinal vascular topology, i.e., how the different vessels are connected to each other, given a single color fundus image. Determining this connectivity is very challenging because vessels cross each other in a 2D image, obscuring their true paths. We validated the usefulness of our extraction method by using it to achieve state-of-the-art results in retinal artery-vein classification. Our proposed approach works as follows. We first segment the retinal vessels using our previously developed state-of-the-art segmentation method. Then, we estimate an initial graph from the extracted vessels and assign the most likely blood flow to each edge. We then use a handful of high-level operations (HLOs) to fix errors in the graph. These HLOs include detaching neighboring nodes, shifting the endpoints of an edge, and reversing the estimated blood flow direction for a branch. We use a novel cost function to find the optimal set of HLO operations for a given graph. Finally, we show that our extracted vascular structure is correct by propagating artery/vein labels along the branches. As our experiments show, our topology-based artery-vein labeling achieved state-of-the-art results on multiple datasets. We also performed several ablation studies to verify the importance of the different components of our proposed method.
Optic Disc Segmentation using Disk-Centered Patch Augmentation
Oct 01, 2021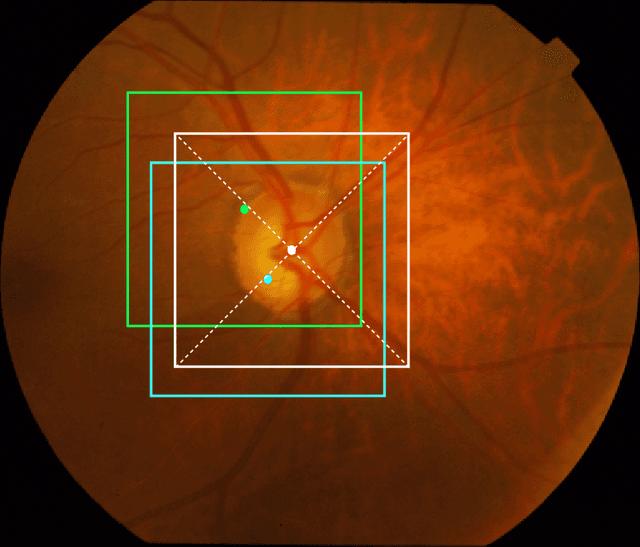
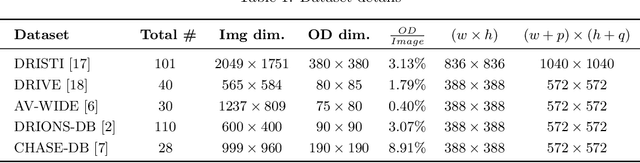
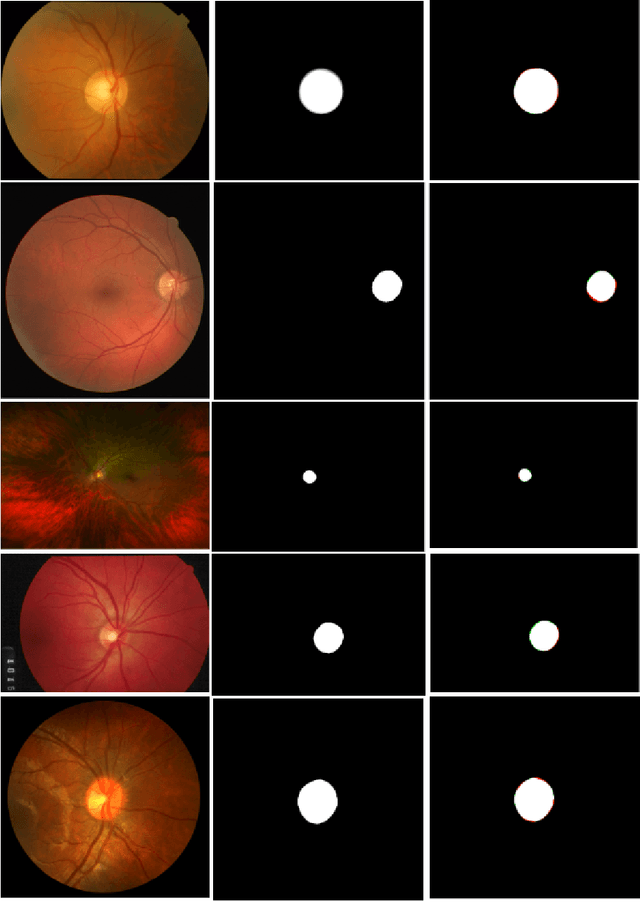
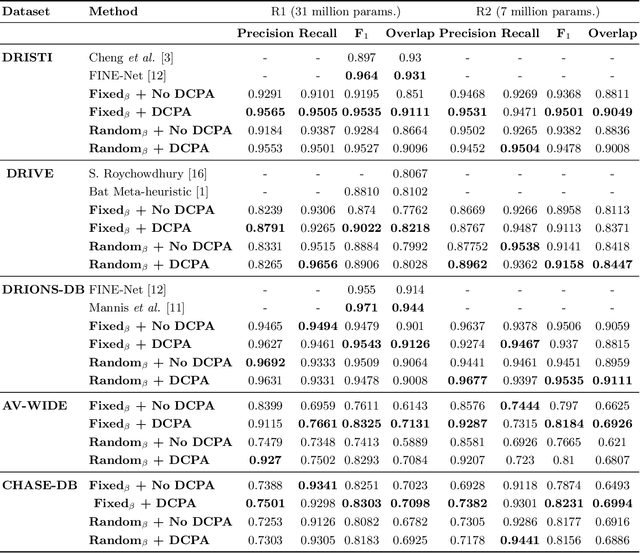
Abstract:The optic disc is a crucial diagnostic feature in the eye since changes to its physiognomy is correlated with the severity of various ocular and cardiovascular diseases. While identifying the bulk of the optic disc in a color fundus image is straightforward, accurately segmenting its boundary at the pixel level is very challenging. In this work, we propose disc-centered patch augmentation (DCPA) -- a simple, yet novel training scheme for deep neural networks -- to address this problem. DCPA achieves state-of-the-art results on full-size images even when using small neural networks, specifically a U-Net with only 7 million parameters as opposed to the original 31 million. In DCPA, we restrict the training data to patches that fully contain the optic nerve. In addition, we also train the network using dynamic cost functions to increase its robustness. We tested DCPA-trained networks on five retinal datasets: DRISTI, DRIONS-DB, DRIVE, AV-WIDE, and CHASE-DB. The first two had available optic disc ground truth, and we manually estimated the ground truth for the latter three. Our approach achieved state-of-the-art F1 and IOU results on four datasets (95 % F1, 91 % IOU on DRISTI; 92 % F1, 84 % IOU on DRIVE; 83 % F1, 71 % IOU on AV-WIDE; 83 % F1, 71 % IOU on CHASEDB) and competitive results on the fifth (95 % F1, 91 % IOU on DRIONS-DB), confirming its generality. Our open-source code and ground-truth annotations are available at: https://github.com/saeidmotevali/fundusdisk
AI Playground: Unreal Engine-based Data Ablation Tool for Deep Learning
Jul 13, 2020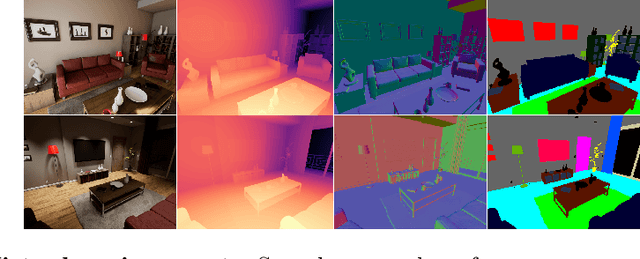
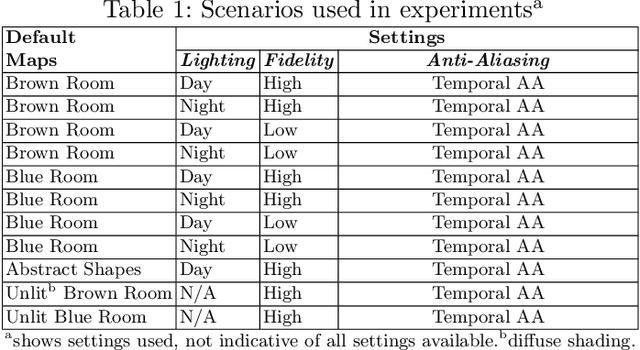
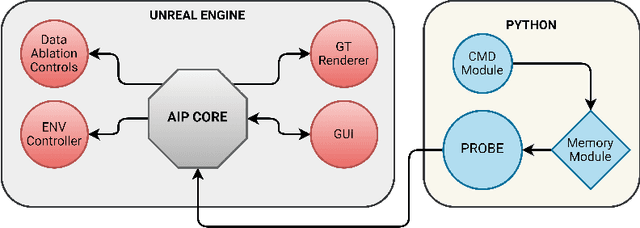

Abstract:Machine learning requires data, but acquiring and labeling real-world data is challenging, expensive, and time-consuming. More importantly, it is nearly impossible to alter real data post-acquisition (e.g., change the illumination of a room), making it very difficult to measure how specific properties of the data affect performance. In this paper, we present AI Playground (AIP), an open-source, Unreal Engine-based tool for generating and labeling virtual image data. With AIP, it is trivial to capture the same image under different conditions (e.g., fidelity, lighting, etc.) and with different ground truths (e.g., depth or surface normal values). AIP is easily extendable and can be used with or without code. To validate our proposed tool, we generated eight datasets of otherwise identical but varying lighting and fidelity conditions. We then trained deep neural networks to predict (1) depth values, (2) surface normals, or (3) object labels and assessed each network's intra- and cross-dataset performance. Among other insights, we verified that sensitivity to different settings is problem-dependent. We confirmed the findings of other studies that segmentation models are very sensitive to fidelity, but we also found that they are just as sensitive to lighting. In contrast, depth and normal estimation models seem to be less sensitive to fidelity or lighting and more sensitive to the structure of the image. Finally, we tested our trained depth-estimation networks on two real-world datasets and obtained results comparable to training on real data alone, confirming that our virtual environments are realistic enough for real-world tasks.
Dynamic Deep Networks for Retinal Vessel Segmentation
Mar 27, 2019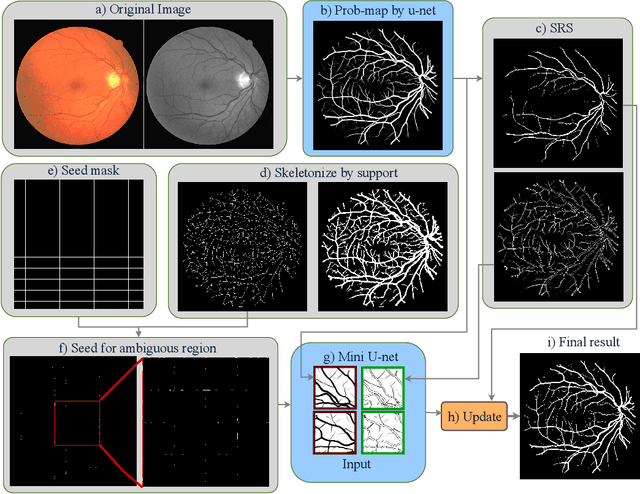
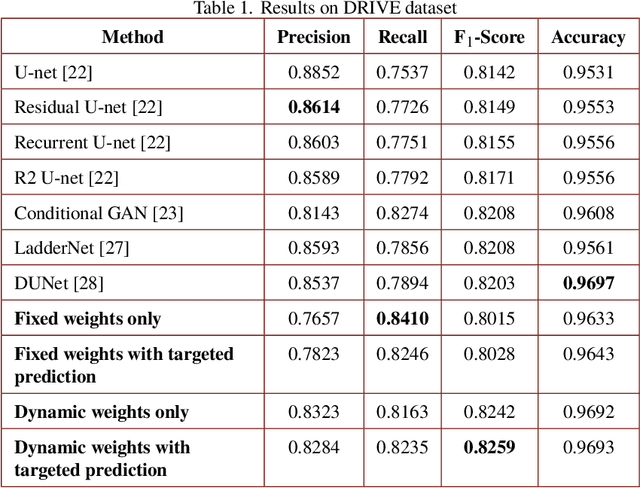
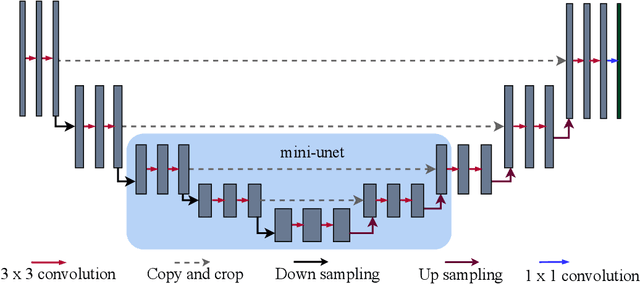
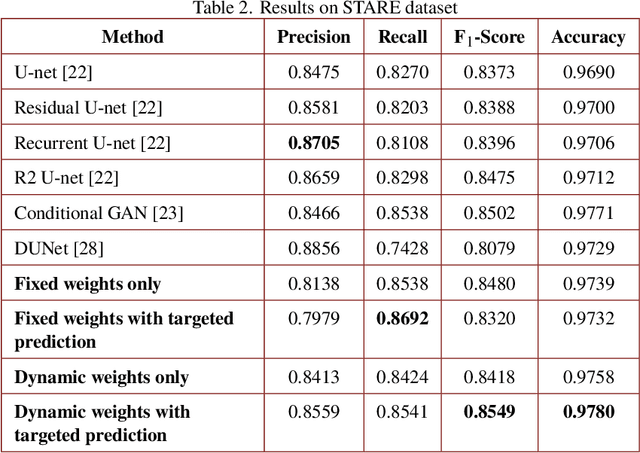
Abstract:Segmenting the retinal vasculature entails a trade-off between how much of the overall vascular structure we identify vs. how precisely we segment individual vessels. In particular, state-of-the-art methods tend to under-segment faint vessels, as well as pixels that lie on the edges of thicker vessels. Thus, they underestimate the width of individual vessels, as well as the ratio of large to small vessels. More generally, many crucial bio-markers---including the artery-vein (AV) ratio, branching angles, number of bifurcation, fractal dimension, tortuosity, vascular length-to-diameter ratio and wall-to-lumen length---require precise measurements of individual vessels. To address this limitation, we propose a novel, stochastic training scheme for deep neural networks that better classifies the faint, ambiguous regions of the image. Our approach relies on two key innovations. First, we train our deep networks with dynamic weights that fluctuate during each training iteration. This stochastic approach forces the network to learn a mapping that robustly balances precision and recall. Second, we decouple the segmentation process into two steps. In the first half of our pipeline, we estimate the likelihood of every pixel and then use these likelihoods to segment pixels that are clearly vessel or background. In the latter part of our pipeline, we use a second network to classify the ambiguous regions in the image. Our proposed method obtained state-of-the-art results on five retinal datasets---DRIVE, STARE, CHASE-DB, AV-WIDE, and VEVIO---by learning a robust balance between false positive and false negative rates. In addition, we are the first to report segmentation results on the AV-WIDE dataset, and we have made the ground-truth annotations for this dataset publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge