Winner-Take-All Computation in Spiking Neural Networks
Paper and Code
Apr 25, 2019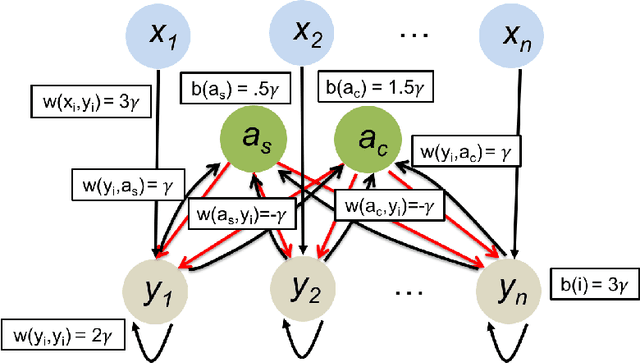
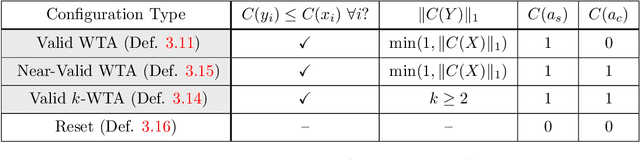
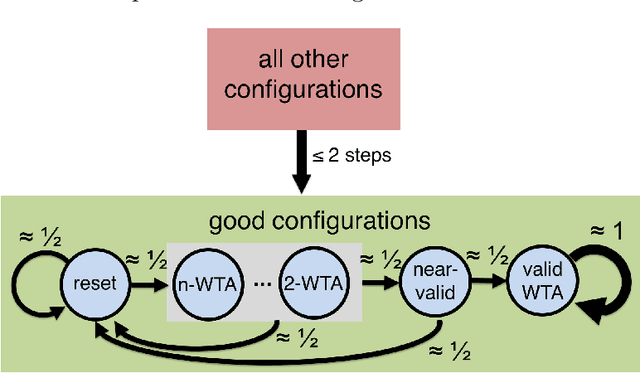
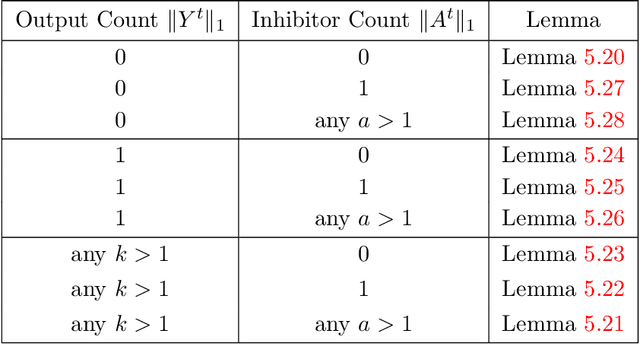
In this work we study biological neural networks from an algorithmic perspective, focusing on understanding tradeoffs between computation time and network complexity. Our goal is to abstract real neural networks in a way that, while not capturing all interesting features, preserves high-level behavior and allows us to make biologically relevant conclusions. Towards this goal, we consider the implementation of algorithmic primitives in a simple yet biologically plausible model of $stochastic\ spiking\ neural\ networks$. In particular, we show how the stochastic behavior of neurons in this model can be leveraged to solve a basic $symmetry-breaking\ task$ in which we are given neurons with identical firing rates and want to select a distinguished one. In computational neuroscience, this is known as the winner-take-all (WTA) problem, and it is believed to serve as a basic building block in many tasks, e.g., learning, pattern recognition, and clustering. We provide efficient constructions of WTA circuits in our stochastic spiking neural network model, as well as lower bounds in terms of the number of auxiliary neurons required to drive convergence to WTA in a given number of steps. These lower bounds demonstrate that our constructions are near-optimal in some cases. This work covers and gives more in-depth proofs of a subset of results originally published in [LMP17a]. It is adapted from the last chapter of C. Musco's Ph.D. thesis [Mus18].
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge