Ulcerative Colitis Mayo Endoscopic Scoring Classification with Active Learning and Generative Data Augmentation
Paper and Code
Nov 10, 2023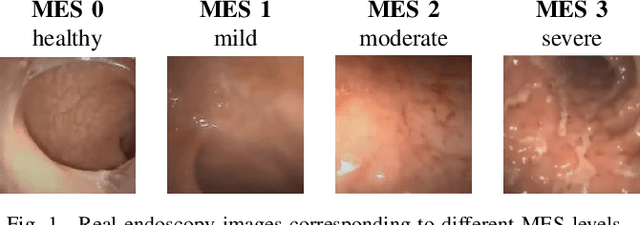
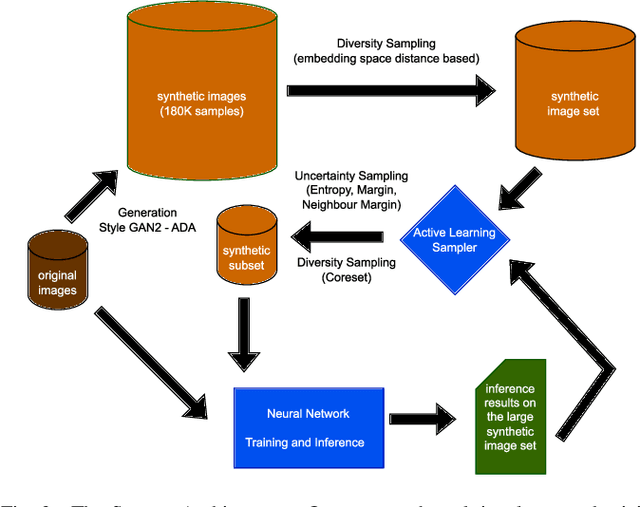
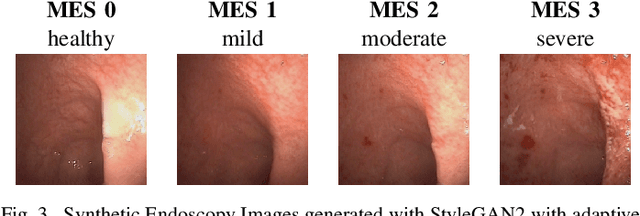
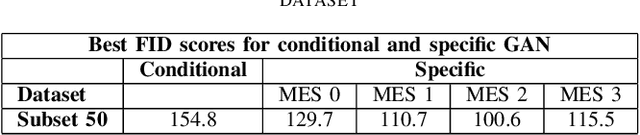
Endoscopic imaging is commonly used to diagnose Ulcerative Colitis (UC) and classify its severity. It has been shown that deep learning based methods are effective in automated analysis of these images and can potentially be used to aid medical doctors. Unleashing the full potential of these methods depends on the availability of large amount of labeled images; however, obtaining and labeling these images are quite challenging. In this paper, we propose a active learning based generative augmentation method. The method involves generating a large number of synthetic samples by training using a small dataset consisting of real endoscopic images. The resulting data pool is narrowed down by using active learning methods to select the most informative samples, which are then used to train a classifier. We demonstrate the effectiveness of our method through experiments on a publicly available endoscopic image dataset. The results show that using synthesized samples in conjunction with active learning leads to improved classification performance compared to using only the original labeled examples and the baseline classification performance of 68.1% increases to 74.5% in terms of Quadratic Weighted Kappa (QWK) Score. Another observation is that, attaining equivalent performance using only real data necessitated three times higher number of images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge