U-learning for Prediction Inference via Combinatory Multi-Subsampling: With Applications to LASSO and Neural Networks
Paper and Code
Jul 22, 2024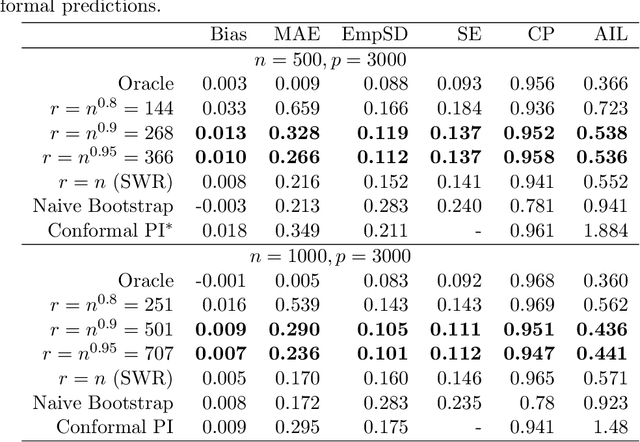
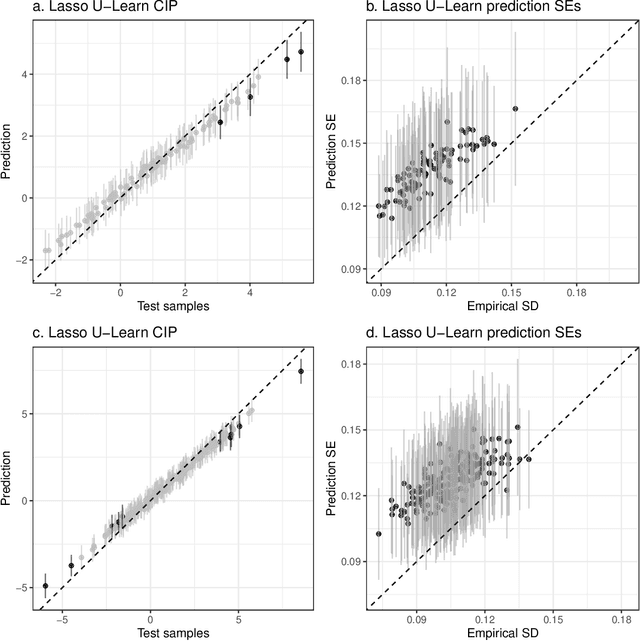
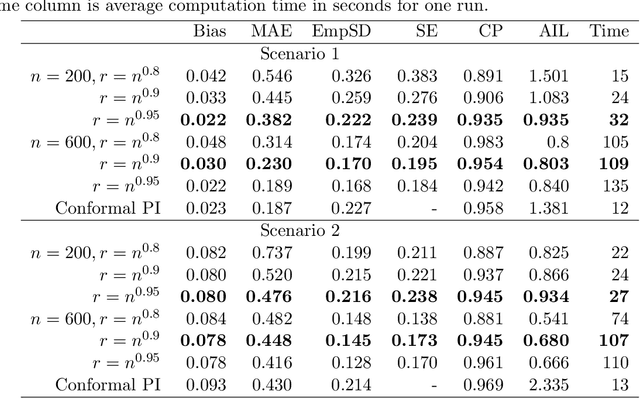
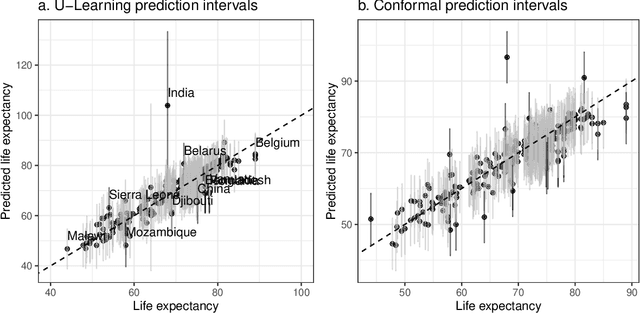
Epigenetic aging clocks play a pivotal role in estimating an individual's biological age through the examination of DNA methylation patterns at numerous CpG (Cytosine-phosphate-Guanine) sites within their genome. However, making valid inferences on predicted epigenetic ages, or more broadly, on predictions derived from high-dimensional inputs, presents challenges. We introduce a novel U-learning approach via combinatory multi-subsampling for making ensemble predictions and constructing confidence intervals for predictions of continuous outcomes when traditional asymptotic methods are not applicable. More specifically, our approach conceptualizes the ensemble estimators within the framework of generalized U-statistics and invokes the H\'ajek projection for deriving the variances of predictions and constructing confidence intervals with valid conditional coverage probabilities. We apply our approach to two commonly used predictive algorithms, Lasso and deep neural networks (DNNs), and illustrate the validity of inferences with extensive numerical studies. We have applied these methods to predict the DNA methylation age (DNAmAge) of patients with various health conditions, aiming to accurately characterize the aging process and potentially guide anti-aging interventions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge