TriResNet: A Deep Triple-stream Residual Network for Histopathology Grading
Paper and Code
Jun 22, 2018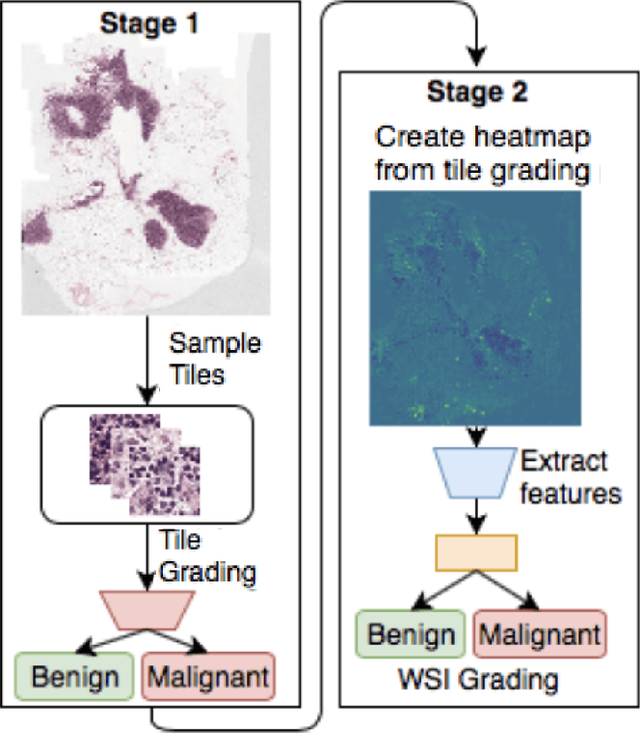
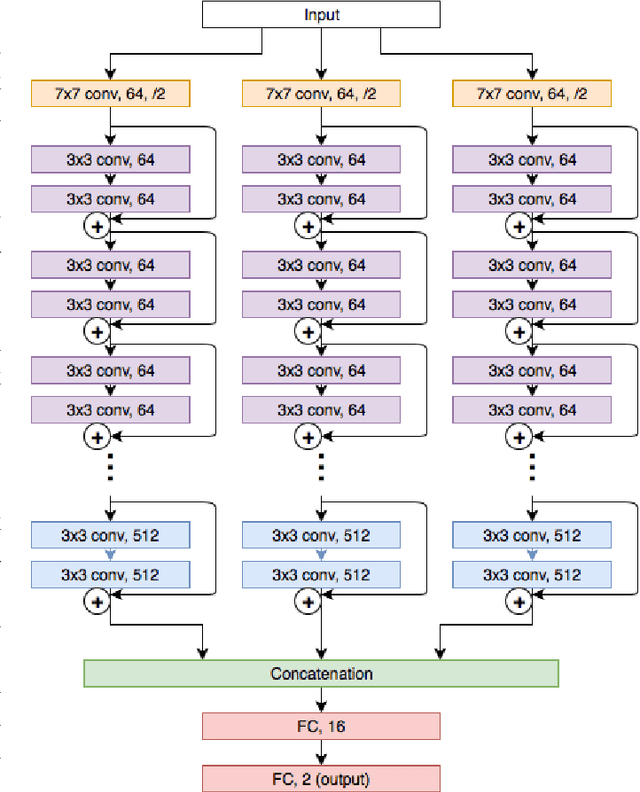
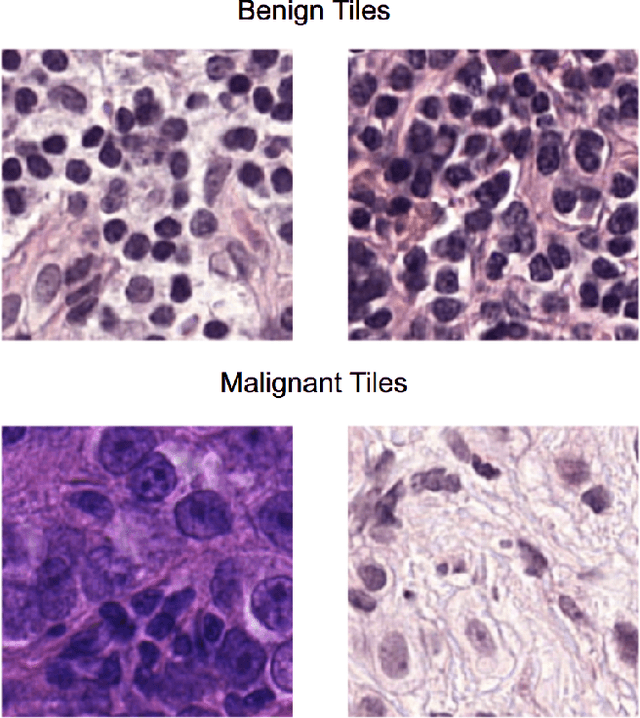
While microscopic analysis of histopathological slides is generally considered as the gold standard method for performing cancer diagnosis and grading, the current method for analysis is extremely time consuming and labour intensive as it requires pathologists to visually inspect tissue samples in a detailed fashion for the presence of cancer. As such, there has been significant recent interest in computer aided diagnosis systems for analysing histopathological slides for cancer grading to aid pathologists to perform cancer diagnosis and grading in a more efficient, accurate, and consistent manner. In this work, we investigate and explore a deep triple-stream residual network (TriResNet) architecture for the purpose of tile-level histopathology grading, which is the critical first step to computer-aided whole-slide histopathology grading. In particular, the design mentality behind the proposed TriResNet network architecture is to facilitate for the learning of a more diverse set of quantitative features to better characterize the complex tissue characteristics found in histopathology samples. Experimental results on two widely-used computer-aided histopathology benchmark datasets (CAMELYON16 dataset and Invasive Ductal Carcinoma (IDC) dataset) demonstrated that the proposed TriResNet network architecture was able to achieve noticeably improved accuracies when compared with two other state-of-the-art deep convolutional neural network architectures. Based on these promising results, the hope is that the proposed TriResNet network architecture could become a useful tool to aiding pathologists increase the consistency, speed, and accuracy of the histopathology grading process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge