Training Generative Adversarial Networks for Optical Property Mapping using Synthetic Image Data
Paper and Code
Mar 15, 2022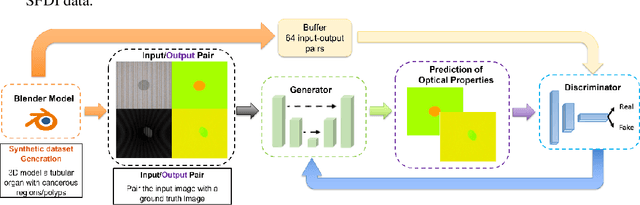
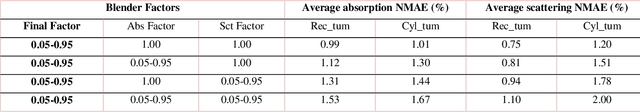
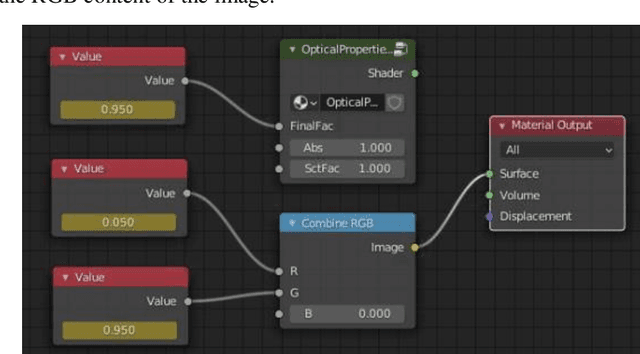
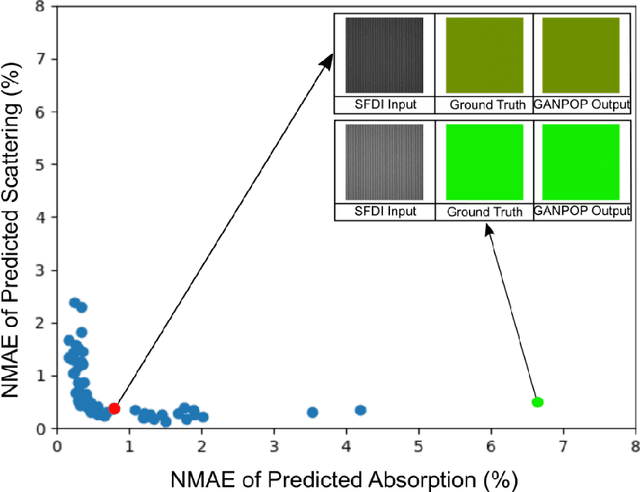
We demonstrate training of a Generative Adversarial Network (GAN) for prediction of optical property maps (scattering and absorption) using spatial frequency domain imaging (SFDI) image data sets generated synthetically with free open-source 3D modelling and rendering software, Blender. The flexibility of Blender is exploited to simulate 3 models with real-life relevance to clinical SFDI of diseased tissue: flat samples, flat samples with spheroidal tumours and cylindrical samples with spheroidal tumours representing imaging inside a tubular organ e.g. the gastro-intestinal tract. In all 3 scenarios we show the GAN provides accurate reconstruction of optical properties from single SFDI images with mean normalised error ranging from 1-1.2% for absorption and 0.7-1.2% for scattering, resulting in visually improved contrast for tumour spheroid structures. This compares favourably with 25% absorption error and 10% scattering error achieved using GANs on experimental SFDI data. However, some of this improvement is due to lower noise and availability of perfect ground truths so we therefore cross-validate our synthetically-trained GAN with a GAN trained on experimental data and observe visually accurate results with error of <40% for absorption and <25% for scattering, due largely to the presence of spatial frequency mismatch artefacts. Our synthetically trained GAN is therefore highly relevant to real experimental samples, but provides significant added benefits of large training datasets, perfect ground-truths and the ability to test realistic imaging geometries, e.g. inside cylinders, for which no conventional single-shot demodulation algorithms exist. In future we expect that application of techniques such as domain adaptation or training on hybrid real-synthetic datasets will create a powerful tool for fast, accurate production of optical property maps from real clinical imaging systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge