Time Scale Network: A Shallow Neural Network For Time Series Data
Paper and Code
Nov 10, 2023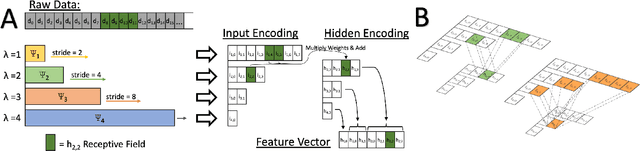
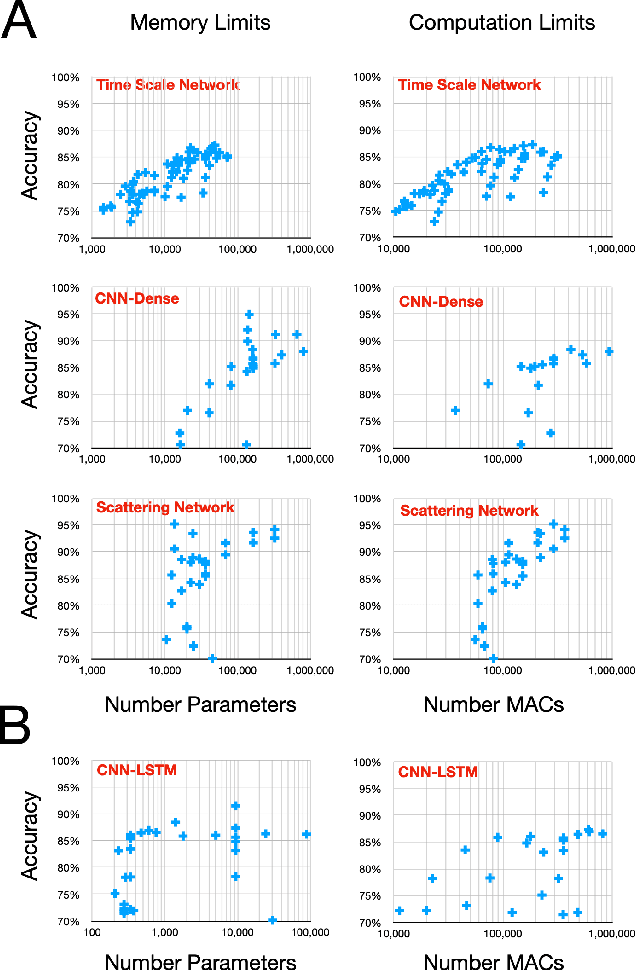
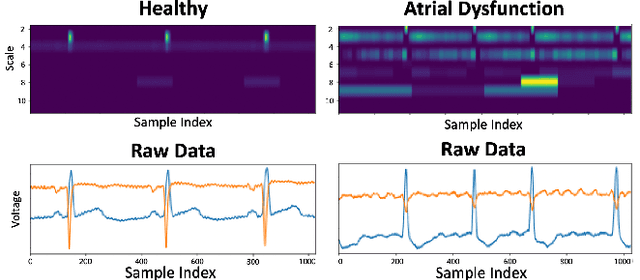
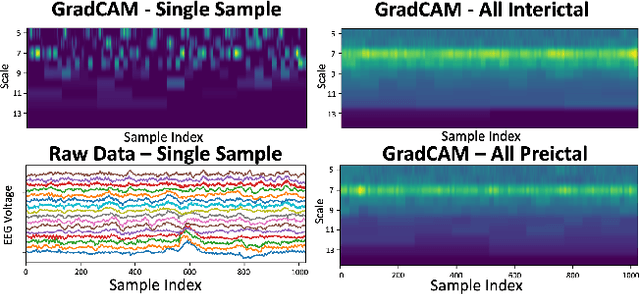
Time series data is often composed of information at multiple time scales, particularly in biomedical data. While numerous deep learning strategies exist to capture this information, many make networks larger, require more data, are more demanding to compute, and are difficult to interpret. This limits their usefulness in real-world applications facing even modest computational or data constraints and can further complicate their translation into practice. We present a minimal, computationally efficient Time Scale Network combining the translation and dilation sequence used in discrete wavelet transforms with traditional convolutional neural networks and back-propagation. The network simultaneously learns features at many time scales for sequence classification with significantly reduced parameters and operations. We demonstrate advantages in Atrial Dysfunction detection including: superior accuracy-per-parameter and accuracy-per-operation, fast training and inference speeds, and visualization and interpretation of learned patterns in atrial dysfunction detection on ECG signals. We also demonstrate impressive performance in seizure prediction using EEG signals. Our network isolated a few time scales that could be strategically selected to achieve 90.9% accuracy using only 1,133 active parameters and consistently converged on pulsatile waveform shapes. This method does not rest on any constraints or assumptions regarding signal content and could be leveraged in any area of time series analysis dealing with signals containing features at many time scales.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge