Syndromic classification of Twitter messages
Paper and Code
Oct 13, 2011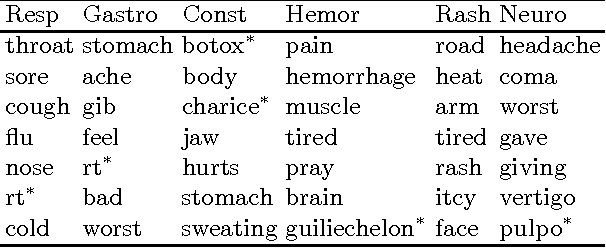
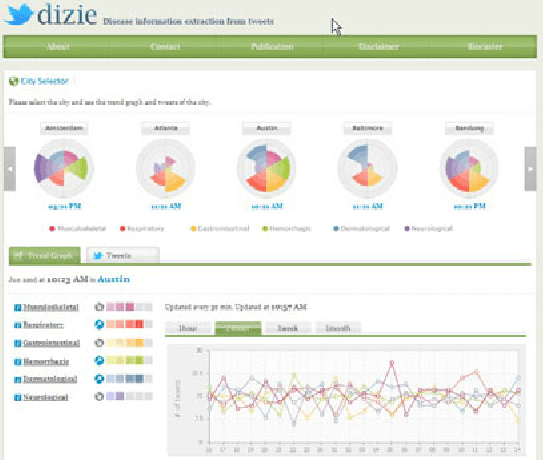
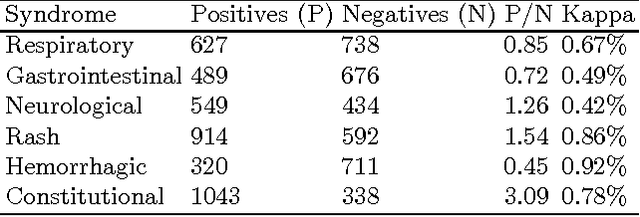
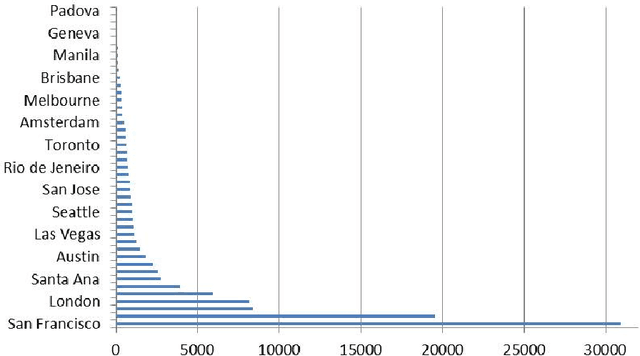
Recent studies have shown strong correlation between social networking data and national influenza rates. We expanded upon this success to develop an automated text mining system that classifies Twitter messages in real time into six syndromic categories based on key terms from a public health ontology. 10-fold cross validation tests were used to compare Naive Bayes (NB) and Support Vector Machine (SVM) models on a corpus of 7431 Twitter messages. SVM performed better than NB on 4 out of 6 syndromes. The best performing classifiers showed moderately strong F1 scores: respiratory = 86.2 (NB); gastrointestinal = 85.4 (SVM polynomial kernel degree 2); neurological = 88.6 (SVM polynomial kernel degree 1); rash = 86.0 (SVM polynomial kernel degree 1); constitutional = 89.3 (SVM polynomial kernel degree 1); hemorrhagic = 89.9 (NB). The resulting classifiers were deployed together with an EARS C2 aberration detection algorithm in an experimental online system.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge