Supervised Parameter Estimation of Neuron Populations from Multiple Firing Events
Paper and Code
Oct 02, 2022
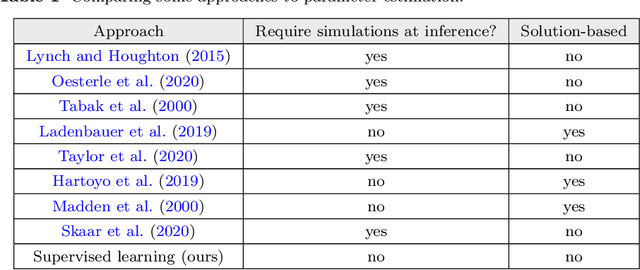
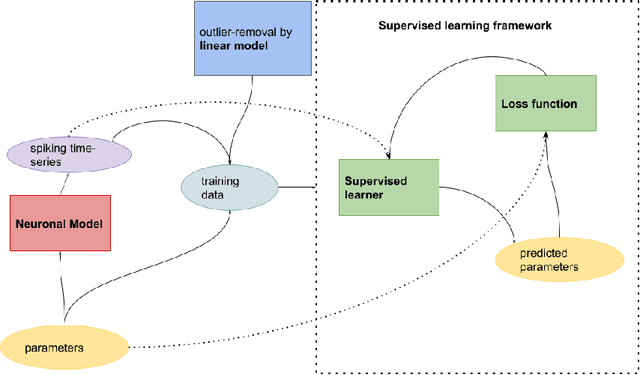

The firing dynamics of biological neurons in mathematical models is often determined by the model's parameters, representing the neurons' underlying properties. The parameter estimation problem seeks to recover those parameters of a single neuron or a neuron population from their responses to external stimuli and interactions between themselves. Most common methods for tackling this problem in the literature use some mechanistic models in conjunction with either a simulation-based or solution-based optimization scheme. In this paper, we study an automatic approach of learning the parameters of neuron populations from a training set consisting of pairs of spiking series and parameter labels via supervised learning. Unlike previous work, this automatic learning does not require additional simulations at inference time nor expert knowledge in deriving an analytical solution or in constructing some approximate models. We simulate many neuronal populations with different parameter settings using a stochastic neuron model. Using that data, we train a variety of supervised machine learning models, including convolutional and deep neural networks, random forest, and support vector regression. We then compare their performance against classical approaches including a genetic search, Bayesian sequential estimation, and a random walk approximate model. The supervised models almost always outperform the classical methods in parameter estimation and spike reconstruction errors, and computation expense. Convolutional neural network, in particular, is the best among all models across all metrics. The supervised models can also generalize to out-of-distribution data to a certain extent.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge