Supervised Learning for Multi-Block Incomplete Data
Paper and Code
Jan 14, 2019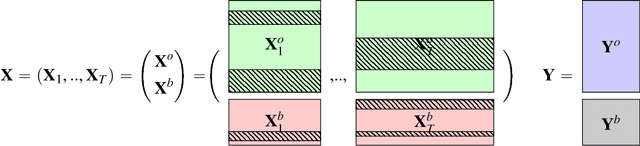
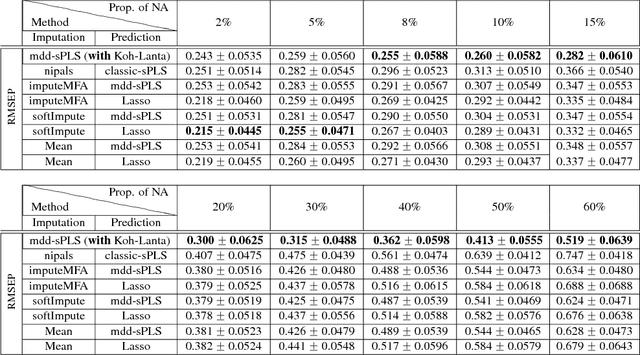
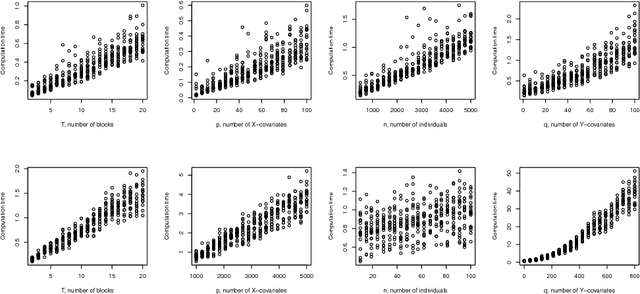
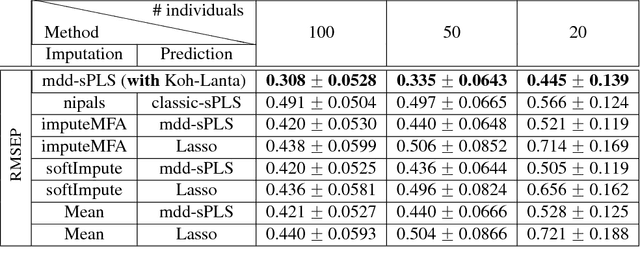
In the supervised high dimensional settings with a large number of variables and a low number of individuals, one objective is to select the relevant variables and thus to reduce the dimension. That subspace selection is often managed with supervised tools. However, some data can be missing, compromising the validity of the sub-space selection. We propose a Partial Least Square (PLS) based method, called Multi-block Data-Driven sparse PLS mdd-sPLS, allowing jointly variable selection and subspace estimation while training and testing missing data imputation through a new algorithm called Koh-Lanta. This method was challenged through simulations against existing methods such as mean imputation, nipals, softImpute and imputeMFA. In the context of supervised analysis of high dimensional data, the proposed method shows the lowest prediction error of the response variables. So far this is the only method combining data imputation and response variable prediction. The superiority of the supervised multi-block mdd-sPLS method increases with the intra-block and inter-block correlations. The application to a real data-set from a rVSV-ZEBOV Ebola vaccine trial revealed interesting and biologically relevant results. The method is implemented in a R-package available on the CRAN and a Python-package available on pypi.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge