Supervised Classification of Flow Cytometric Samples via the Joint Clustering and Matching (JCM) Procedure
Paper and Code
Nov 11, 2014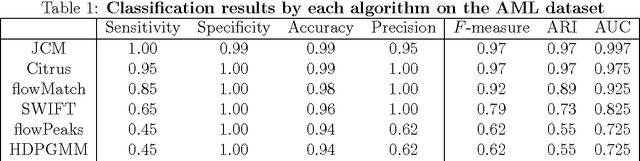
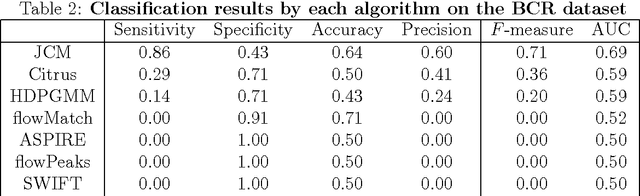
We consider the use of the Joint Clustering and Matching (JCM) procedure for the supervised classification of a flow cytometric sample with respect to a number of predefined classes of such samples. The JCM procedure has been proposed as a method for the unsupervised classification of cells within a sample into a number of clusters and in the case of multiple samples, the matching of these clusters across the samples. The two tasks of clustering and matching of the clusters are performed simultaneously within the JCM framework. In this paper, we consider the case where there is a number of distinct classes of samples whose class of origin is known, and the problem is to classify a new sample of unknown class of origin to one of these predefined classes. For example, the different classes might correspond to the types of a particular disease or to the various health outcomes of a patient subsequent to a course of treatment. We show and demonstrate on some real datasets how the JCM procedure can be used to carry out this supervised classification task. A mixture distribution is used to model the distribution of the expressions of a fixed set of markers for each cell in a sample with the components in the mixture model corresponding to the various populations of cells in the composition of the sample. For each class of samples, a class template is formed by the adoption of random-effects terms to model the inter-sample variation within a class. The classification of a new unclassified sample is undertaken by assigning the unclassified sample to the class that minimizes the Kullback-Leibler distance between its fitted mixture density and each class density provided by the class templates.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge