Stochastic Optimization of 3D Non-Cartesian Sampling Trajectory (SNOPY)
Paper and Code
Sep 22, 2022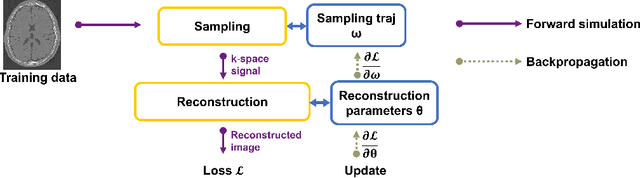
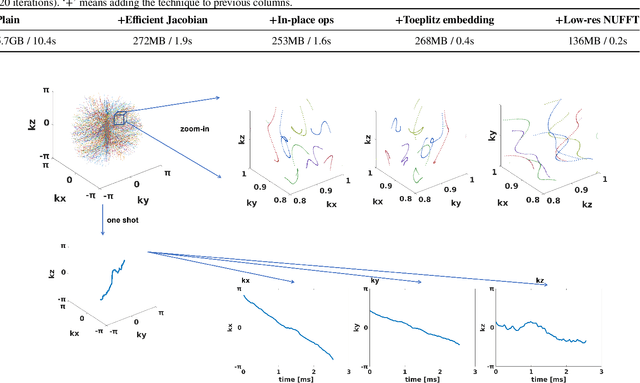
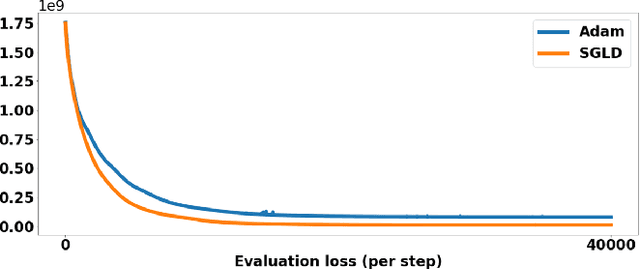

Optimizing 3D k-space sampling trajectories for efficient MRI is important yet challenging. This work proposes a generalized framework for optimizing 3D non-Cartesian sampling patterns via data-driven optimization. We built a differentiable MRI system model to enable gradient-based methods for sampling trajectory optimization. By combining training losses, the algorithm can simultaneously optimize multiple properties of sampling patterns, including image quality, hardware constraints (maximum slew rate and gradient strength), reduced peripheral nerve stimulation (PNS), and parameter-weighted contrast. The proposed method can either optimize the gradient waveform (spline-based freeform optimization) or optimize properties of given sampling trajectories (such as the rotation angle of radial trajectories). Notably, the method optimizes sampling trajectories synergistically with either model-based or learning-based reconstruction methods. We proposed several strategies to alleviate the severe non-convexity and huge computation demand posed by the high-dimensional optimization. The corresponding code is organized as an open-source, easy-to-use toolbox. We applied the optimized trajectory to multiple applications including structural and functional imaging. In the simulation studies, the reconstruction PSNR of a 3D kooshball trajectory was increased by 4 dB with SNOPY optimization. In the prospective studies, by optimizing the rotation angles of a stack-of-stars (SOS) trajectory, SNOPY improved the PSNR by 1.4dB compared to the best empirical method. Optimizing the gradient waveform of a rotational EPI trajectory improved subjects' rating of the PNS effect from 'strong' to 'mild.' In short, SNOPY provides an efficient data-driven and optimization-based method to tailor non-Cartesian sampling trajectories.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge