SparGE: Sparse Coding-based Patient Similarity Learning via Low-rank Constraints and Graph Embedding
Paper and Code
Feb 03, 2022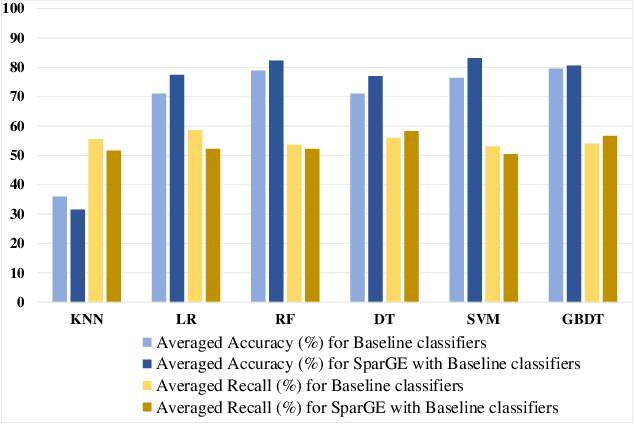
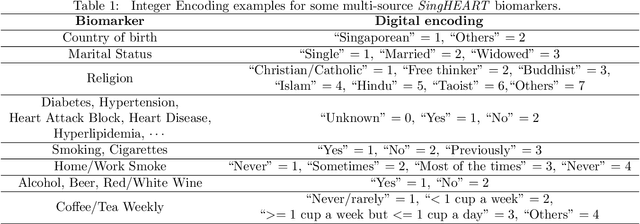
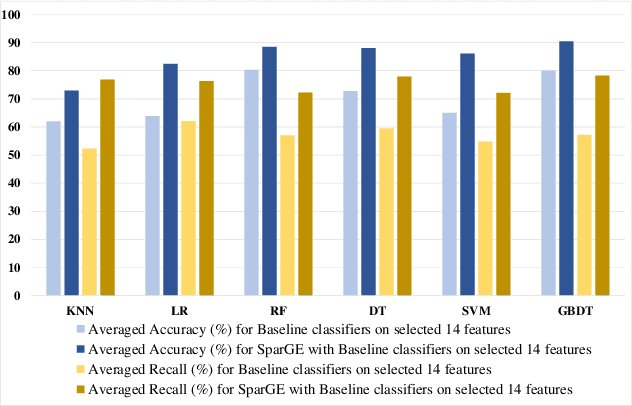

Patient similarity assessment (PSA) is pivotal to evidence-based and personalized medicine, enabled by analyzing the increasingly available electronic health records (EHRs). However, machine learning approaches for PSA has to deal with inherent data deficiencies of EHRs, namely missing values, noise, and small sample sizes. In this work, an end-to-end discriminative learning framework, called SparGE, is proposed to address these data challenges of EHR for PSA. SparGE measures similarity by jointly sparse coding and graph embedding. First, we use low-rank constrained sparse coding to identify and calculate weight for similar patients, while denoising against missing values. Then, graph embedding on sparse representations is adopted to measure the similarity between patient pairs via preserving local relationships defined by distances. Finally, a global cost function is constructed to optimize related parameters. Experimental results on two private and public real-world healthcare datasets, namely SingHEART and MIMIC-III, show that the proposed SparGE significantly outperforms other machine learning patient similarity methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge