Solving Label Variation in Scientific Information Extraction via Multi-Task Learning
Paper and Code
Dec 25, 2023
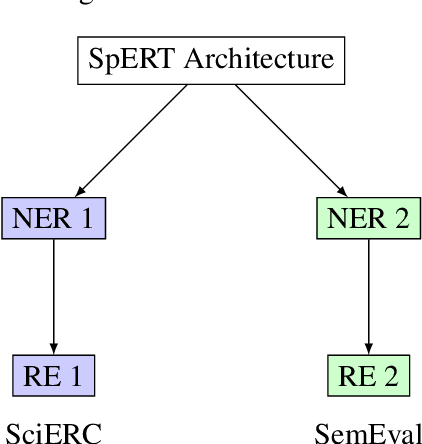


Scientific Information Extraction (ScientificIE) is a critical task that involves the identification of scientific entities and their relationships. The complexity of this task is compounded by the necessity for domain-specific knowledge and the limited availability of annotated data. Two of the most popular datasets for ScientificIE are SemEval-2018 Task-7 and SciERC. They have overlapping samples and differ in their annotation schemes, which leads to conflicts. In this study, we first introduced a novel approach based on multi-task learning to address label variations. We then proposed a soft labeling technique that converts inconsistent labels into probabilistic distributions. The experimental results demonstrated that the proposed method can enhance the model robustness to label noise and improve the end-to-end performance in both ScientificIE tasks. The analysis revealed that label variations can be particularly effective in handling ambiguous instances. Furthermore, the richness of the information captured by label variations can potentially reduce data size requirements. The findings highlight the importance of releasing variation labels and promote future research on other tasks in other domains. Overall, this study demonstrates the effectiveness of multi-task learning and the potential of label variations to enhance the performance of ScientificIE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge