Simulation assisted machine learning
Paper and Code
Feb 20, 2018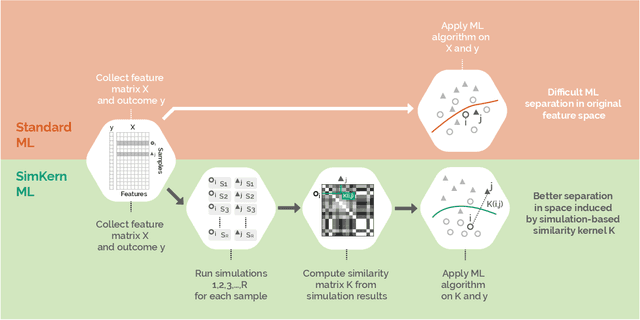
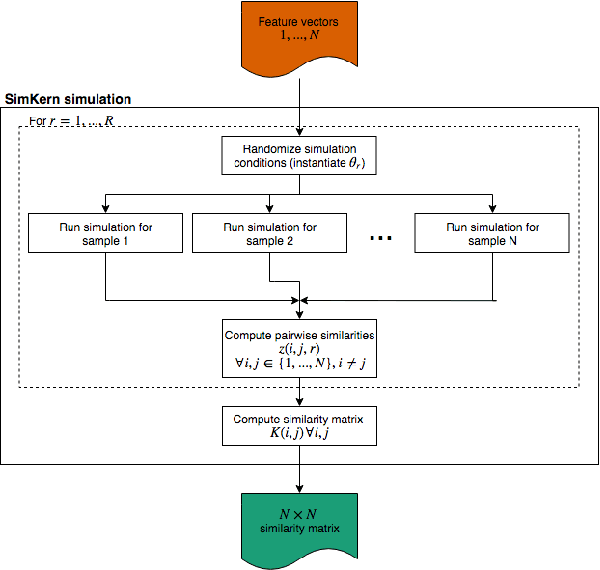
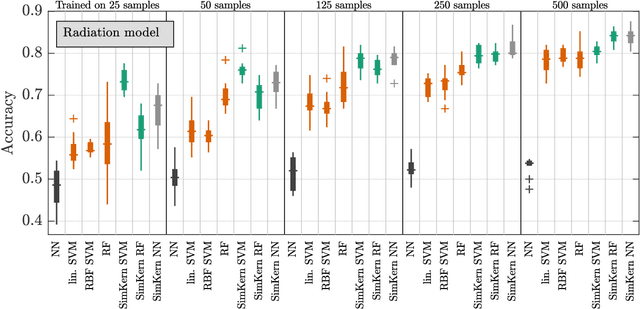
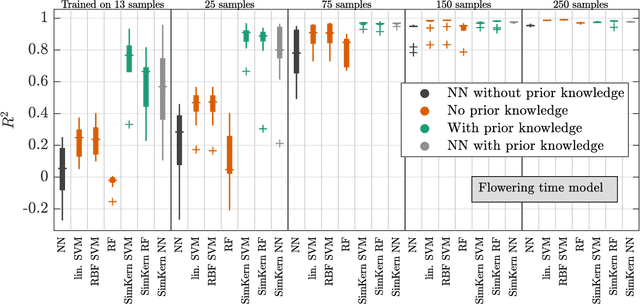
Predicting how a proposed cancer treatment will affect a given tumor can be cast as a machine learning problem, but the complexity of biological systems, the number of potentially relevant genomic and clinical features, and the lack of very large scale patient data repositories make this a unique challenge. "Pure data" approaches to this problem are underpowered to detect combinatorially complex interactions and are bound to uncover false correlations despite statistical precautions taken (1). To investigate this setting, we propose a method to integrate simulations, a strong form of prior knowledge, into machine learning, a combination which to date has been largely unexplored. The results of multiple simulations (under various uncertainty scenarios) are used to compute similarity measures between every pair of samples: sample pairs are given a high similarity score if they behave similarly under a wide range of simulation parameters. These similarity values, rather than the original high dimensional feature data, are used to train kernelized machine learning algorithms such as support vector machines, thus handling the curse-of-dimensionality that typically affects genomic machine learning. Using four synthetic datasets of complex systems--three biological models and one network flow optimization model--we demonstrate that when the number of training samples is small compared to the number of features, the simulation kernel approach dominates over no-prior-knowledge methods. In addition to biology and medicine, this approach should be applicable to other disciplines, such as weather forecasting, financial markets, and agricultural management, where predictive models are sought and informative yet approximate simulations are available. The Python SimKern software, the models (in MATLAB, Octave, and R), and the datasets are made freely available at https://github.com/davidcraft/SimKern .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge