Self-supervised Learning for Acoustic Few-Shot Classification
Paper and Code
Sep 15, 2024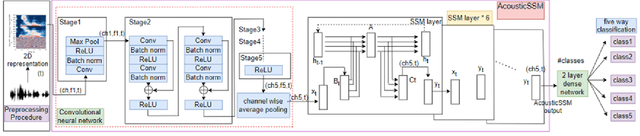
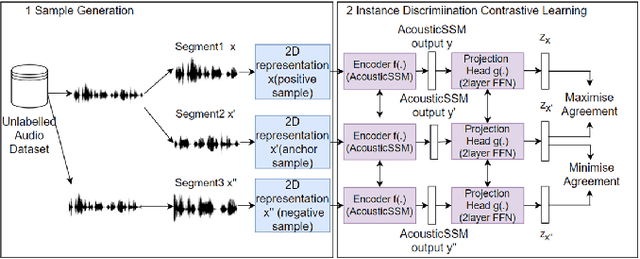
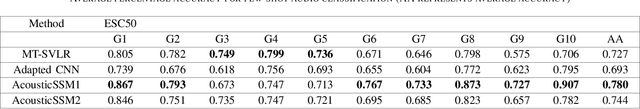
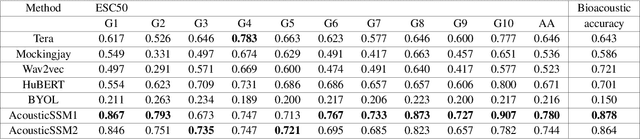
Labelled data are limited and self-supervised learning is one of the most important approaches for reducing labelling requirements. While it has been extensively explored in the image domain, it has so far not received the same amount of attention in the acoustic domain. Yet, reducing labelling is a key requirement for many acoustic applications. Specifically in bioacoustic, there are rarely sufficient labels for fully supervised learning available. This has led to the widespread use of acoustic recognisers that have been pre-trained on unrelated data for bioacoustic tasks. We posit that training on the actual task data and combining self-supervised pre-training with few-shot classification is a superior approach that has the ability to deliver high accuracy even when only a few labels are available. To this end, we introduce and evaluate a new architecture that combines CNN-based preprocessing with feature extraction based on state space models (SSMs). This combination is motivated by the fact that CNN-based networks alone struggle to capture temporal information effectively, which is crucial for classifying acoustic signals. SSMs, specifically S4 and Mamba, on the other hand, have been shown to have an excellent ability to capture long-range dependencies in sequence data. We pre-train this architecture using contrastive learning on the actual task data and subsequent fine-tuning with an extremely small amount of labelled data. We evaluate the performance of this proposed architecture for ($n$-shot, $n$-class) classification on standard benchmarks as well as real-world data. Our evaluation shows that it outperforms state-of-the-art architectures on the few-shot classification problem.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge