Scalable, Axiomatic Explanations of Deep Alzheimer's Diagnosis from Heterogeneous Data
Paper and Code
Jul 13, 2021
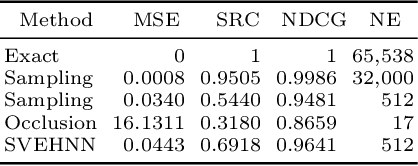
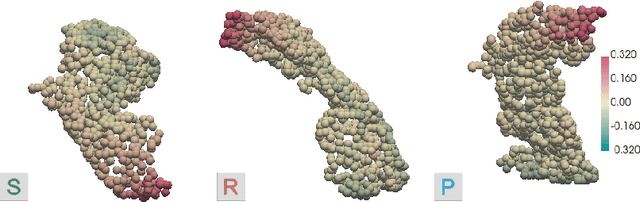

Deep Neural Networks (DNNs) have an enormous potential to learn from complex biomedical data. In particular, DNNs have been used to seamlessly fuse heterogeneous information from neuroanatomy, genetics, biomarkers, and neuropsychological tests for highly accurate Alzheimer's disease diagnosis. On the other hand, their black-box nature is still a barrier for the adoption of such a system in the clinic, where interpretability is absolutely essential. We propose Shapley Value Explanation of Heterogeneous Neural Networks (SVEHNN) for explaining the Alzheimer's diagnosis made by a DNN from the 3D point cloud of the neuroanatomy and tabular biomarkers. Our explanations are based on the Shapley value, which is the unique method that satisfies all fundamental axioms for local explanations previously established in the literature. Thus, SVEHNN has many desirable characteristics that previous work on interpretability for medical decision making is lacking. To avoid the exponential time complexity of the Shapley value, we propose to transform a given DNN into a Lightweight Probabilistic Deep Network without re-training, thus achieving a complexity only quadratic in the number of features. In our experiments on synthetic and real data, we show that we can closely approximate the exact Shapley value with a dramatically reduced runtime and can reveal the hidden knowledge the network has learned from the data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge