SAFE setup for generative molecular design
Paper and Code
Oct 26, 2024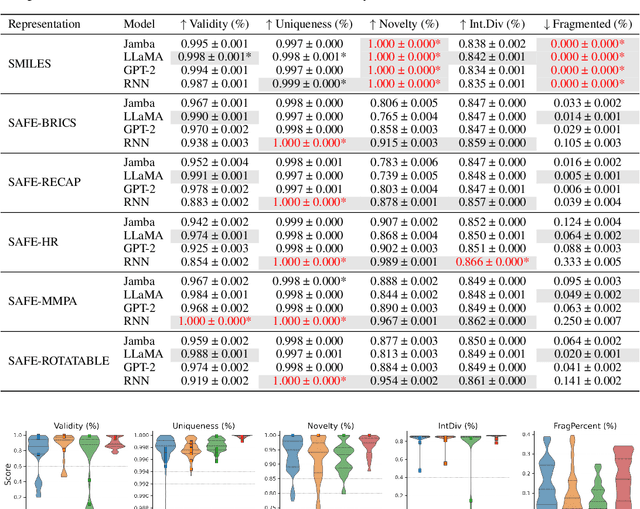
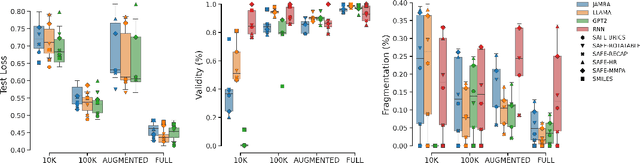
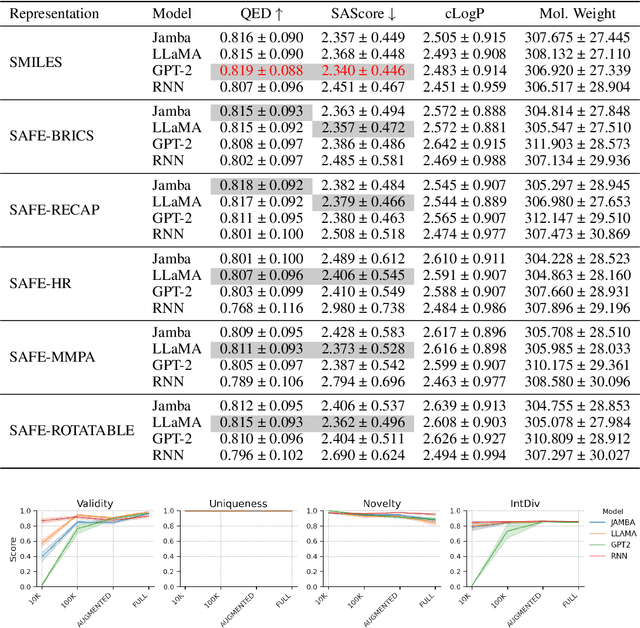
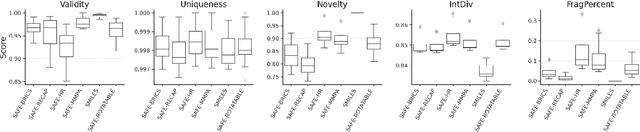
SMILES-based molecular generative models have been pivotal in drug design but face challenges in fragment-constrained tasks. To address this, the Sequential Attachment-based Fragment Embedding (SAFE) representation was recently introduced as an alternative that streamlines those tasks. In this study, we investigate the optimal setups for training SAFE generative models, focusing on dataset size, data augmentation through randomization, model architecture, and bond disconnection algorithms. We found that larger, more diverse datasets improve performance, with the LLaMA architecture using Rotary Positional Embedding proving most robust. SAFE-based models also consistently outperform SMILES-based approaches in scaffold decoration and linker design, particularly with BRICS decomposition yielding the best results. These insights highlight key factors that significantly impact the efficacy of SAFE-based generative models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge