Robust Gaussian Graphical Modeling with the Trimmed Graphical Lasso
Paper and Code
Oct 28, 2015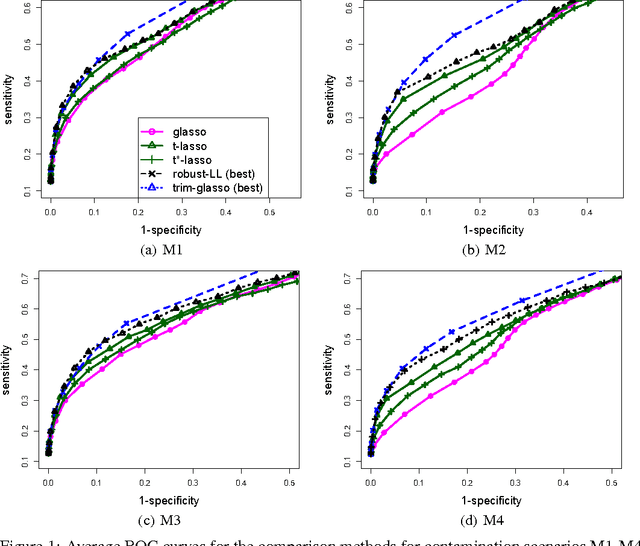
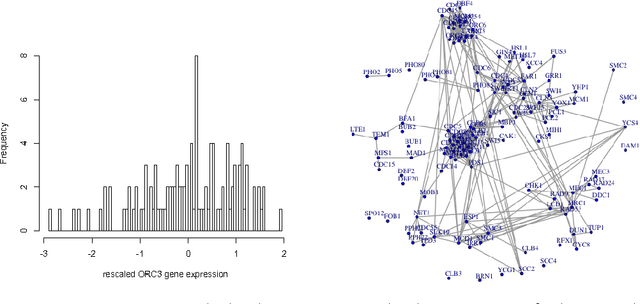
Gaussian Graphical Models (GGMs) are popular tools for studying network structures. However, many modern applications such as gene network discovery and social interactions analysis often involve high-dimensional noisy data with outliers or heavier tails than the Gaussian distribution. In this paper, we propose the Trimmed Graphical Lasso for robust estimation of sparse GGMs. Our method guards against outliers by an implicit trimming mechanism akin to the popular Least Trimmed Squares method used for linear regression. We provide a rigorous statistical analysis of our estimator in the high-dimensional setting. In contrast, existing approaches for robust sparse GGMs estimation lack statistical guarantees. Our theoretical results are complemented by experiments on simulated and real gene expression data which further demonstrate the value of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge