Prediction and optimization of NaV1.7 inhibitors based on machine learning methods
Paper and Code
Nov 29, 2019
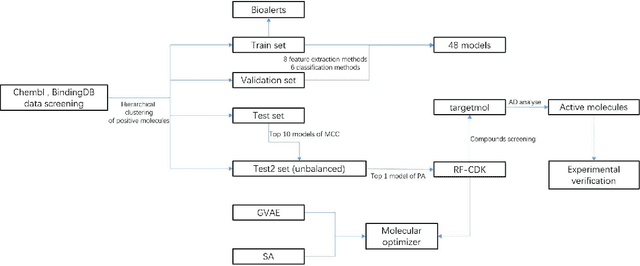


Objectives NaV1.7 is a key target related to pain. This study focused on predicting and optimizing inhibitors of NaV1.7 using machine learning methods, and using patch-clamp methods to validate them at the cellular level. Materials and Methods We used Chembl, BindingDB, and data from the literature to establish classification models for inhibitors. The imbalanced data set test2 was used to screen the best-performing model to screen commercial compound libraries, and whole-cell voltage-clamp was used to validate inhibitors. We propose a molecular group optimization method using a combination of Grammer Variational Autoencoder, classification model, and simulated annealing algorithm. Results and Conclusion We get the model RF-CDK that performs best in the imbalanced data set. Of the three compounds that may have inhibitory effects, Nortriptyline has been experimentally verified. In the molecular optimization method, the best result of the optimization results of CHEMBL2325245 is MS = 1.052, PROB = 0.527, SA = 2.587, QED = 0.462. 40 molecules located in the applicability domain of RF-CDK were used for optimization, among which 34 molecules gave larger MS values.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge