Organ-aware Multi-scale Medical Image Segmentation Using Text Prompt Engineering
Paper and Code
Mar 18, 2025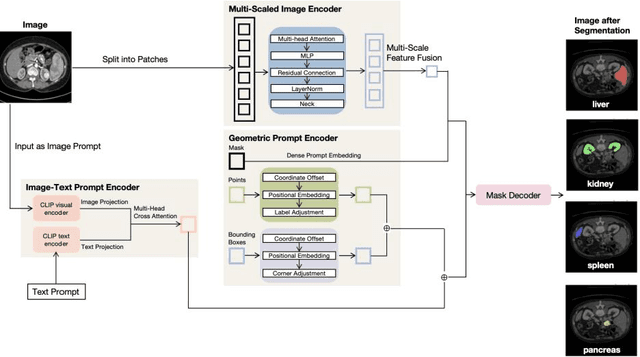
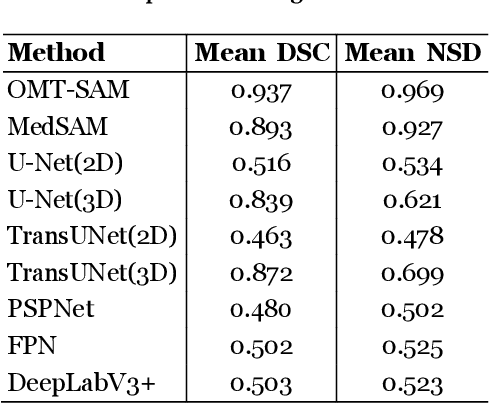
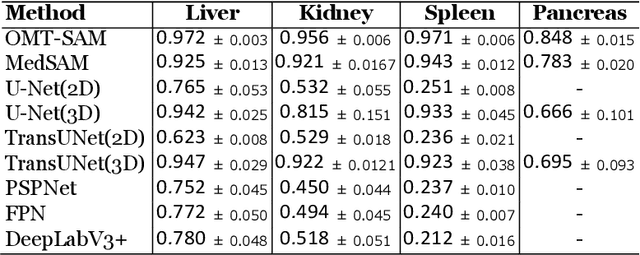
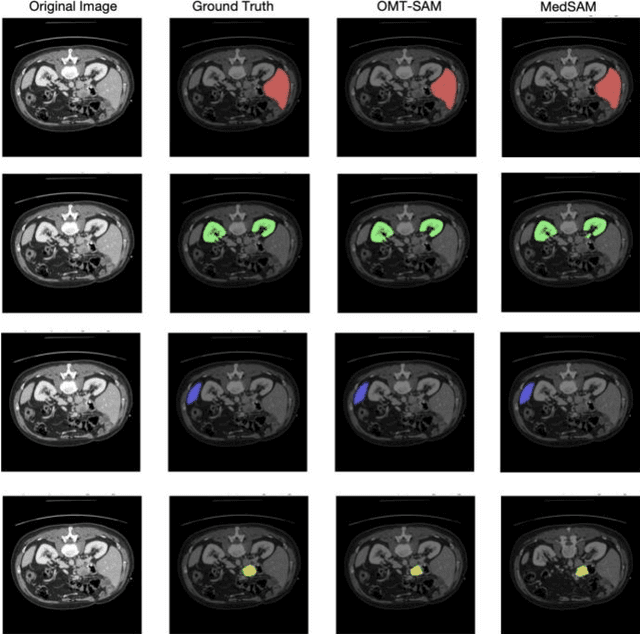
Accurate segmentation is essential for effective treatment planning and disease monitoring. Existing medical image segmentation methods predominantly rely on uni-modal visual inputs, such as images or videos, requiring labor-intensive manual annotations. Additionally, medical imaging techniques capture multiple intertwined organs within a single scan, further complicating segmentation accuracy. To address these challenges, MedSAM, a large-scale medical segmentation model based on the Segment Anything Model (SAM), was developed to enhance segmentation accuracy by integrating image features with user-provided prompts. While MedSAM has demonstrated strong performance across various medical segmentation tasks, it primarily relies on geometric prompts (e.g., points and bounding boxes) and lacks support for text-based prompts, which could help specify subtle or ambiguous anatomical structures. To overcome these limitations, we propose the Organ-aware Multi-scale Text-guided Medical Image Segmentation Model (OMT-SAM) for multi-organ segmentation. Our approach introduces CLIP encoders as a novel image-text prompt encoder, operating with the geometric prompt encoder to provide informative contextual guidance. We pair descriptive textual prompts with corresponding images, processing them through pre-trained CLIP encoders and a cross-attention mechanism to generate fused image-text embeddings. Additionally, we extract multi-scale visual features from MedSAM, capturing fine-grained anatomical details at different levels of granularity. We evaluate OMT-SAM on the FLARE 2021 dataset, benchmarking its performance against existing segmentation methods. Empirical results demonstrate that OMT-SAM achieves a mean Dice Similarity Coefficient of 0.937, outperforming MedSAM (0.893) and other segmentation models, highlighting its superior capability in handling complex medical image segmentation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge