optimalFlow: Optimal-transport approach to flow cytometry gating and population matching
Paper and Code
Jul 18, 2019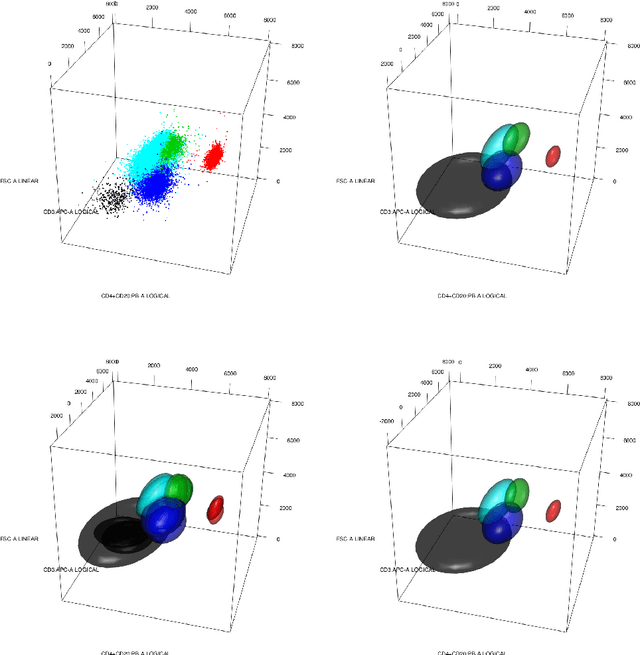
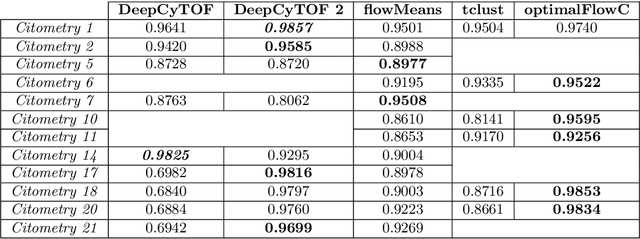
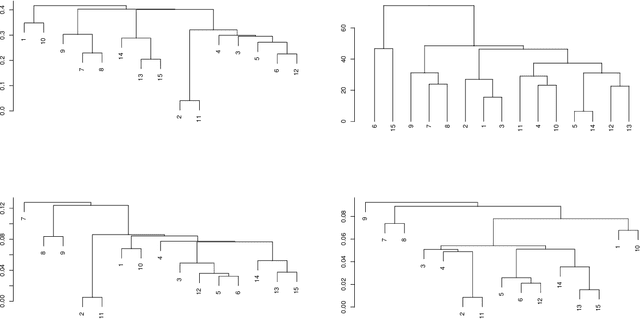
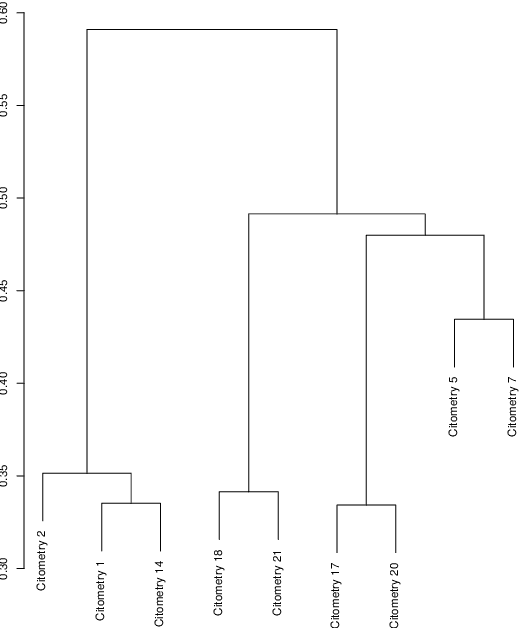
Data used in Flow Cytometry present pronounced variability due to biological and technical reasons. Biological variability is a well known phenomenon produced by measurements on different individuals, with different characteristics such as age, sex, etc... The use of different settings for measurement, the variation of the conditions during experiments or the different types of flow cytometers are some of the technical sources of variability. This high variability makes difficult the use of supervised machine learning for identification of cell populations. We propose optimalFlowTemplates, based on a similarity distance and Wasserstein barycenters, which clusterizes cytometries and produces prototype cytometries for the different groups. We show that supervised learning restricted to the new groups performs better than the same techniques applied to the whole collection. We also present optimalFlowClassification, which uses a database of gated cytometries and optimalFlowTemplates to assign cell types to a new cytometry. We show that this procedure can outperform state of the art techniques in the proposed datasets. Our code and data are freely available as R packages at https://github.com/HristoInouzhe/optimalFlow and https://github.com/HristoInouzhe/optimalFlowData.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge