Note: Variational Encoding of Protein Dynamics Benefits from Maximizing Latent Autocorrelation
Paper and Code
Mar 17, 2018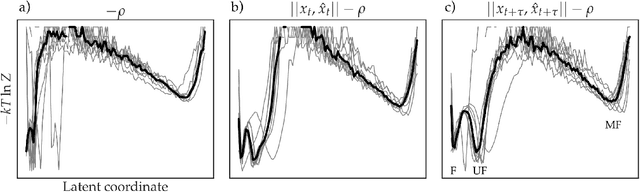
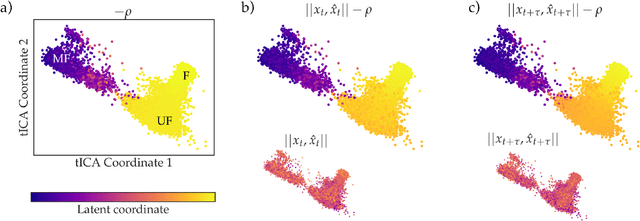
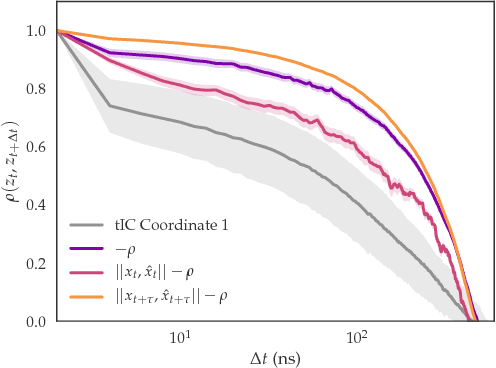
As deep Variational Auto-Encoder (VAE) frameworks become more widely used for modeling biomolecular simulation data, we emphasize the capability of the VAE architecture to concurrently maximize the timescale of the latent space while inferring a reduced coordinate, which assists in finding slow processes as according to the variational approach to conformational dynamics. We additionally provide evidence that the VDE framework (Hern\'andez et al., 2017), which uses this autocorrelation loss along with a time-lagged reconstruction loss, obtains a variationally optimized latent coordinate in comparison with related loss functions. We thus recommend leveraging the autocorrelation of the latent space while training neural network models of biomolecular simulation data to better represent slow processes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge