Multistep Automated Data Labelling Procedure (MADLaP) for Thyroid Nodules on Ultrasound: An Artificial Intelligence Approach for Automating Image Annotation
Paper and Code
Jun 28, 2022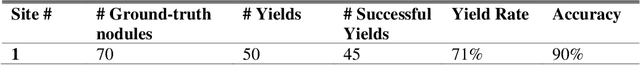
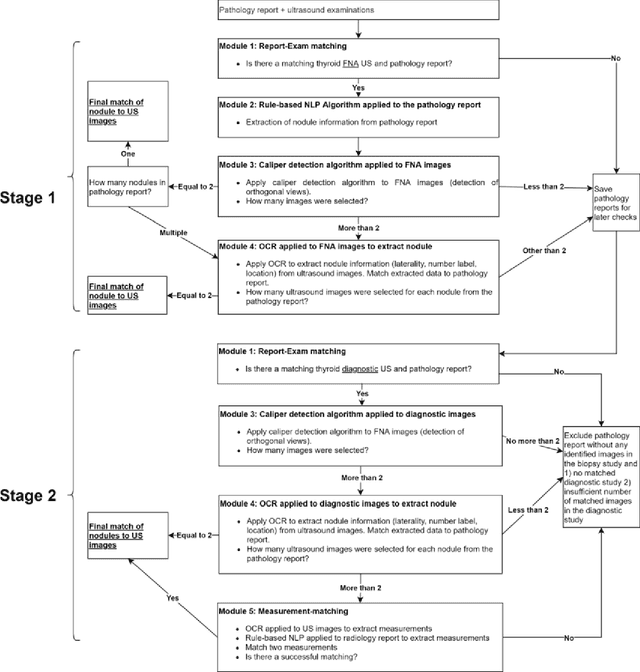
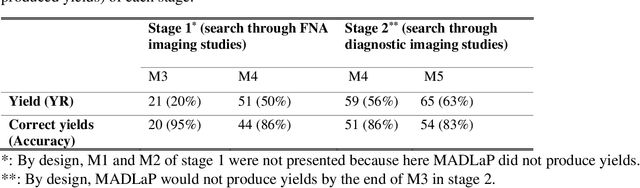

Machine learning (ML) for diagnosis of thyroid nodules on ultrasound is an active area of research. However, ML tools require large, well-labelled datasets, the curation of which is time-consuming and labor-intensive. The purpose of our study was to develop and test a deep-learning-based tool to facilitate and automate the data annotation process for thyroid nodules; we named our tool Multistep Automated Data Labelling Procedure (MADLaP). MADLaP was designed to take multiple inputs included pathology reports, ultrasound images, and radiology reports. Using multiple step-wise modules including rule-based natural language processing, deep-learning-based imaging segmentation, and optical character recognition, MADLaP automatically identified images of a specific thyroid nodule and correctly assigned a pathology label. The model was developed using a training set of 378 patients across our health system and tested on a separate set of 93 patients. Ground truths for both sets were selected by an experienced radiologist. Performance metrics including yield (how many labeled images the model produced) and accuracy (percentage correct) were measured using the test set. MADLaP achieved a yield of 63% and an accuracy of 83%. The yield progressively increased as the input data moved through each module, while accuracy peaked part way through. Error analysis showed that inputs from certain examination sites had lower accuracy (40%) than the other sites (90%, 100%). MADLaP successfully created curated datasets of labeled ultrasound images of thyroid nodules. While accurate, the relatively suboptimal yield of MADLaP exposed some challenges when trying to automatically label radiology images from heterogeneous sources. The complex task of image curation and annotation could be automated, allowing for enrichment of larger datasets for use in machine learning development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge