MolMix: A Simple Yet Effective Baseline for Multimodal Molecular Representation Learning
Paper and Code
Oct 10, 2024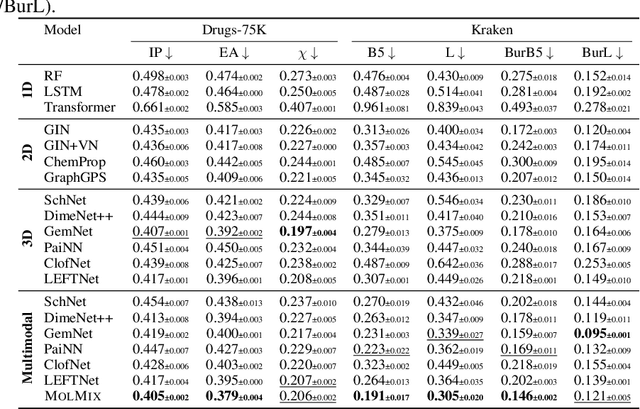
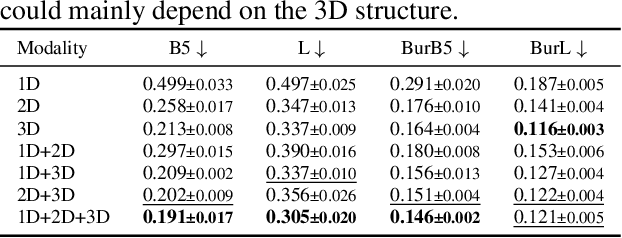
In this work, we propose a simple transformer-based baseline for multimodal molecular representation learning, integrating three distinct modalities: SMILES strings, 2D graph representations, and 3D conformers of molecules. A key aspect of our approach is the aggregation of 3D conformers, allowing the model to account for the fact that molecules can adopt multiple conformations-an important factor for accurate molecular representation. The tokens for each modality are extracted using modality-specific encoders: a transformer for SMILES strings, a message-passing neural network for 2D graphs, and an equivariant neural network for 3D conformers. The flexibility and modularity of this framework enable easy adaptation and replacement of these encoders, making the model highly versatile for different molecular tasks. The extracted tokens are then combined into a unified multimodal sequence, which is processed by a downstream transformer for prediction tasks. To efficiently scale our model for large multimodal datasets, we utilize Flash Attention 2 and bfloat16 precision. Despite its simplicity, our approach achieves state-of-the-art results across multiple datasets, demonstrating its effectiveness as a strong baseline for multimodal molecular representation learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge