Modeling Diverse Chemical Reactions for Single-step Retrosynthesis via Discrete Latent Variables
Paper and Code
Aug 10, 2022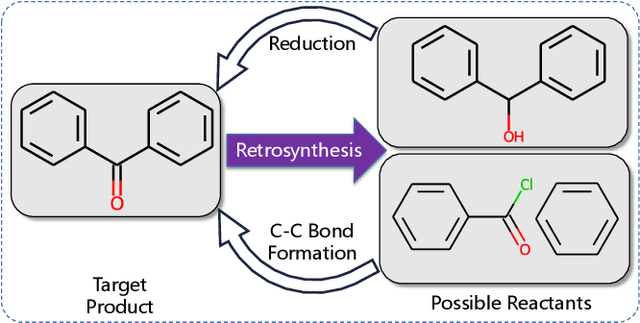
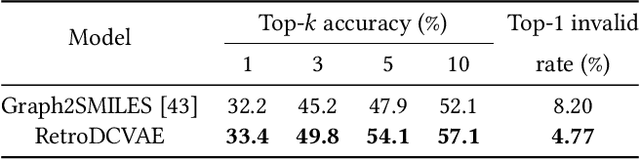
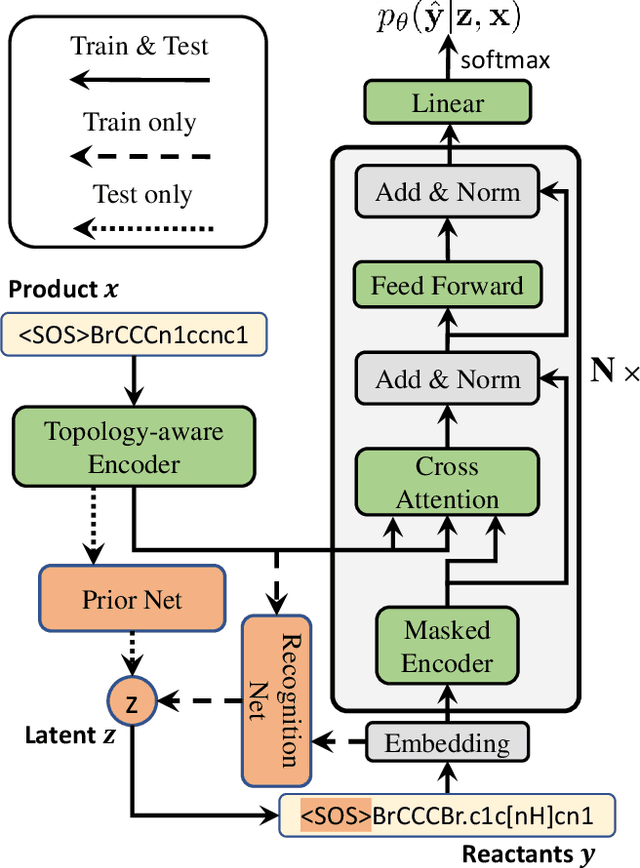
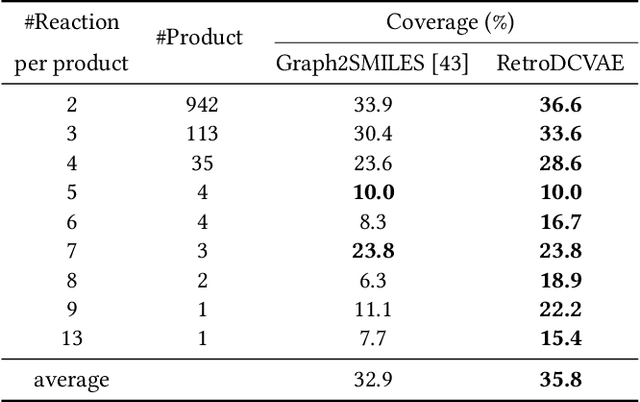
Single-step retrosynthesis is the cornerstone of retrosynthesis planning, which is a crucial task for computer-aided drug discovery. The goal of single-step retrosynthesis is to identify the possible reactants that lead to the synthesis of the target product in one reaction. By representing organic molecules as canonical strings, existing sequence-based retrosynthetic methods treat the product-to-reactant retrosynthesis as a sequence-to-sequence translation problem. However, most of them struggle to identify diverse chemical reactions for a desired product due to the deterministic inference, which contradicts the fact that many compounds can be synthesized through various reaction types with different sets of reactants. In this work, we aim to increase reaction diversity and generate various reactants using discrete latent variables. We propose a novel sequence-based approach, namely RetroDVCAE, which incorporates conditional variational autoencoders into single-step retrosynthesis and associates discrete latent variables with the generation process. Specifically, RetroDVCAE uses the Gumbel-Softmax distribution to approximate the categorical distribution over potential reactions and generates multiple sets of reactants with the variational decoder. Experiments demonstrate that RetroDVCAE outperforms state-of-the-art baselines on both benchmark dataset and homemade dataset. Both quantitative and qualitative results show that RetroDVCAE can model the multi-modal distribution over reaction types and produce diverse reactant candidates.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge