MassFormer: Tandem Mass Spectrum Prediction with Graph Transformers
Paper and Code
Nov 15, 2021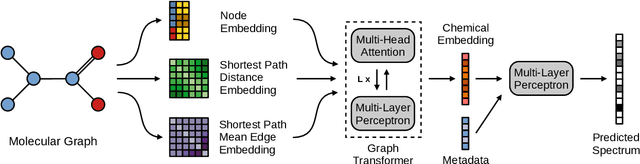
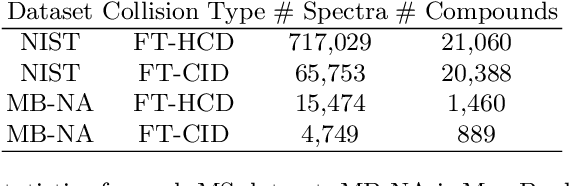
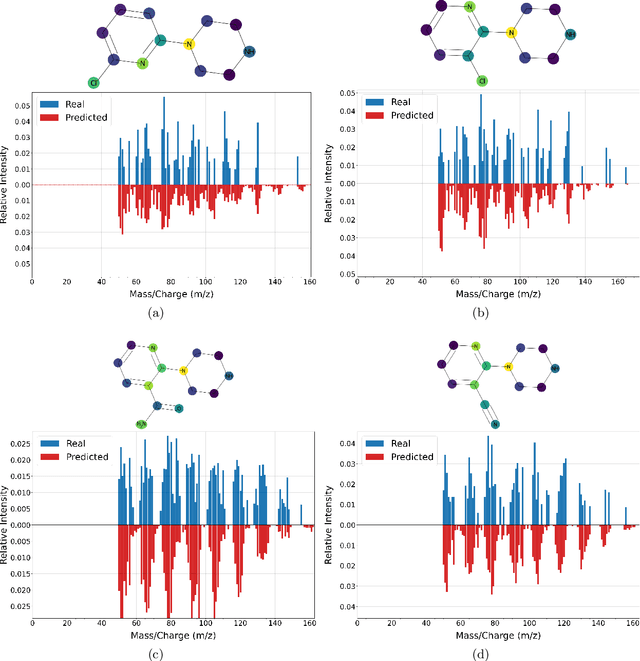
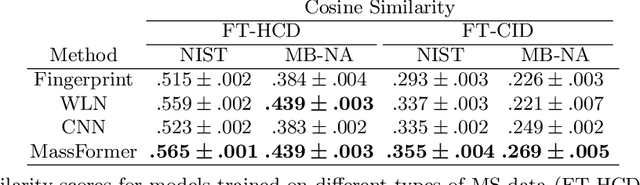
Mass spectrometry is a key tool in the study of small molecules, playing an important role in metabolomics, drug discovery, and environmental chemistry. Tandem mass spectra capture fragmentation patterns that provide key structural information about a molecule and help with its identification. Practitioners often rely on spectral library searches to match unknown spectra with known compounds. However, such search-based methods are limited by availability of reference experimental data. In this work we show that graph transformers can be used to accurately predict tandem mass spectra. Our model, MassFormer, outperforms competing deep learning approaches for spectrum prediction, and includes an interpretable attention mechanism to help explain predictions. We demonstrate that our model can be used to improve reference library coverage on a synthetic molecule identification task. Through quantitative analysis and visual inspection, we verify that our model recovers prior knowledge about the effect of collision energy on the generated spectrum. We evaluate our model on different types of mass spectra from two independent MS datasets and show that its performance generalizes. Code available at github.com/Roestlab/massformer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge