Linked Deep Gaussian Process Emulation for Model Networks
Paper and Code
Jun 02, 2023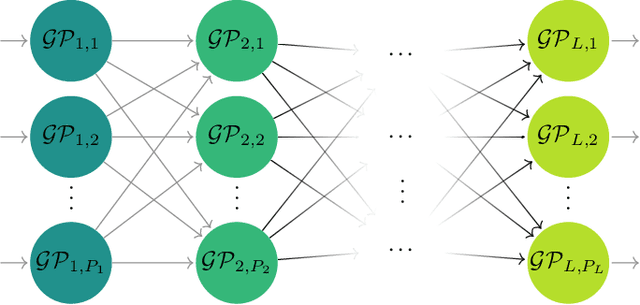
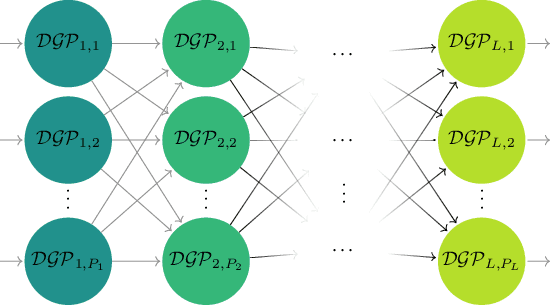
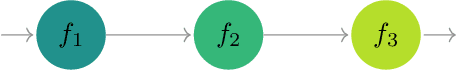
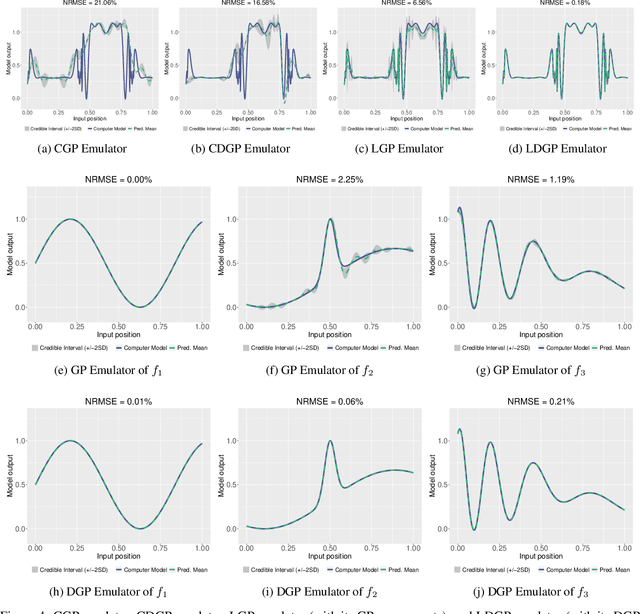
Modern scientific problems are often multi-disciplinary and require integration of computer models from different disciplines, each with distinct functional complexities, programming environments, and computation times. Linked Gaussian process (LGP) emulation tackles this challenge through a divide-and-conquer strategy that integrates Gaussian process emulators of the individual computer models in a network. However, the required stationarity of the component Gaussian process emulators within the LGP framework limits its applicability in many real-world applications. In this work, we conceptualize a network of computer models as a deep Gaussian process with partial exposure of its hidden layers. We develop a method for inference for these partially exposed deep networks that retains a key strength of the LGP framework, whereby each model can be emulated separately using a DGP and then linked together. We show in both synthetic and empirical examples that our linked deep Gaussian process emulators exhibit significantly better predictive performance than standard LGP emulators in terms of accuracy and uncertainty quantification. They also outperform single DGPs fitted to the network as a whole because they are able to integrate information from the partially exposed hidden layers. Our methods are implemented in an R package $\texttt{dgpsi}$ that is freely available on CRAN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge