Leveraging Sequence Embedding and Convolutional Neural Network for Protein Function Prediction
Paper and Code
Dec 01, 2021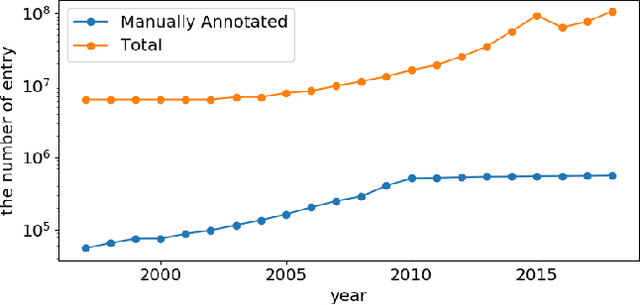
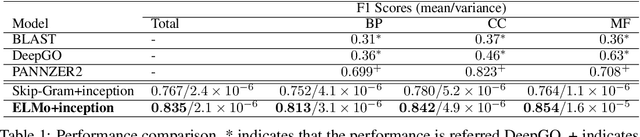
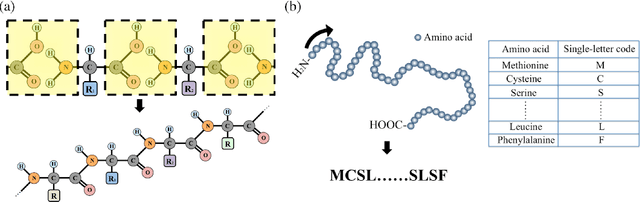
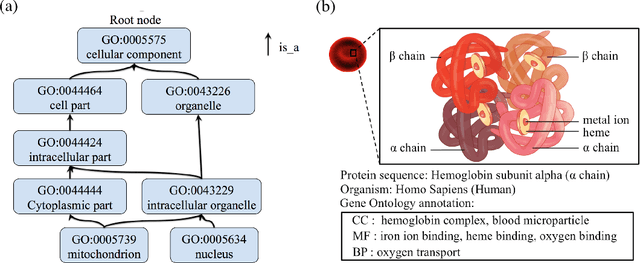
The capability of accurate prediction of protein functions and properties is essential in the biotechnology industry, e.g. drug development and artificial protein synthesis, etc. The main challenges of protein function prediction are the large label space and the lack of labeled training data. Our method leverages unsupervised sequence embedding and the success of deep convolutional neural network to overcome these challenges. In contrast, most of the existing methods delete the rare protein functions to reduce the label space. Furthermore, some existing methods require additional bio-information (e.g., the 3-dimensional structure of the proteins) which is difficult to be determined in biochemical experiments. Our proposed method significantly outperforms the other methods on the publicly available benchmark using only protein sequences as input. This allows the process of identifying protein functions to be sped up.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge