Latent Tree Decomposition Parsers for AMR-to-Text Generation
Paper and Code
Sep 01, 2021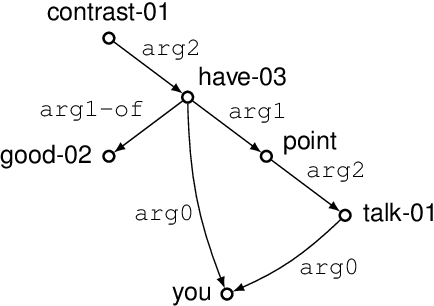
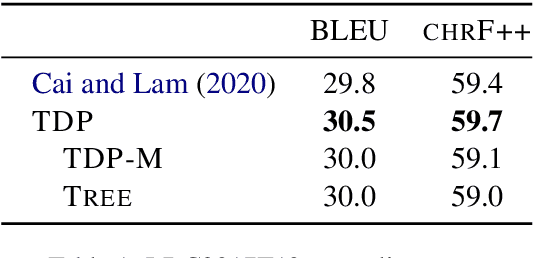
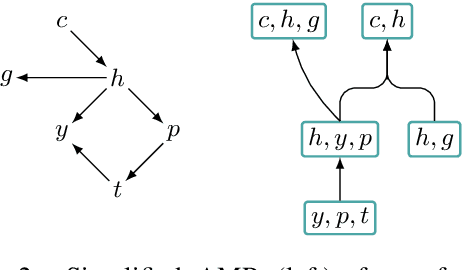

Graph encoders in AMR-to-text generation models often rely on neighborhood convolutions or global vertex attention. While these approaches apply to general graphs, AMRs may be amenable to encoders that target their tree-like structure. By clustering edges into a hierarchy, a tree decomposition summarizes graph structure. Our model encodes a derivation forest of tree decompositions and extracts an expected tree. From tree node embeddings, it builds graph edge features used in vertex attention of the graph encoder. Encoding TD forests instead of shortest-pairwise paths in a self-attentive baseline raises BLEU by 0.7 and chrF++ by 0.3. The forest encoder also surpasses a convolutional baseline for molecular property prediction by 1.92% ROC-AUC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge