Large-scale protein-protein post-translational modification extraction with distant supervision and confidence calibrated BioBERT
Paper and Code
Jan 06, 2022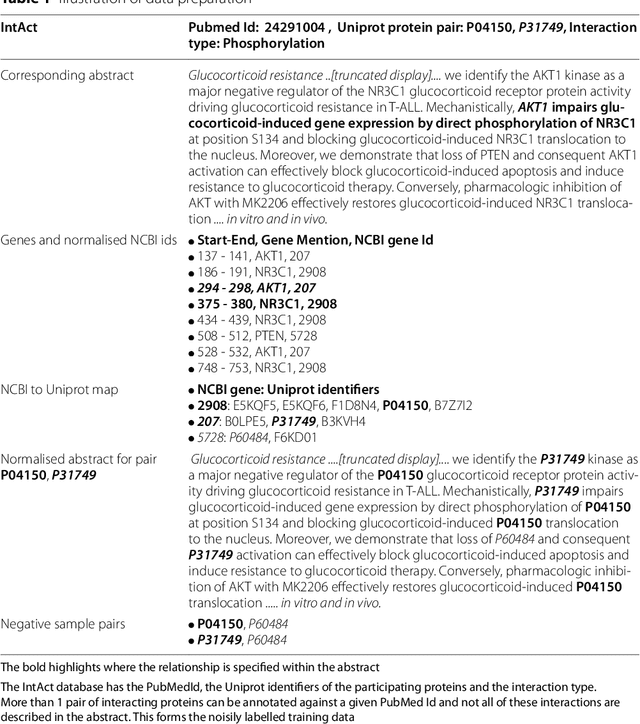


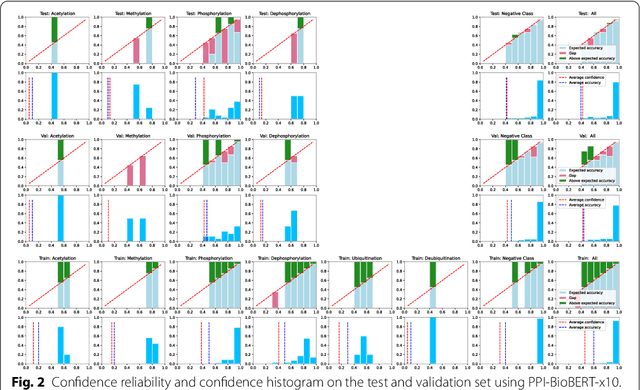
Protein-protein interactions (PPIs) are critical to normal cellular function and are related to many disease pathways. However, only 4% of PPIs are annotated with PTMs in biological knowledge databases such as IntAct, mainly performed through manual curation, which is neither time nor cost-effective. We use the IntAct PPI database to create a distant supervised dataset annotated with interacting protein pairs, their corresponding PTM type, and associated abstracts from the PubMed database. We train an ensemble of BioBERT models - dubbed PPI-BioBERT-x10 to improve confidence calibration. We extend the use of ensemble average confidence approach with confidence variation to counteract the effects of class imbalance to extract high confidence predictions. The PPI-BioBERT-x10 model evaluated on the test set resulted in a modest F1-micro 41.3 (P =5 8.1, R = 32.1). However, by combining high confidence and low variation to identify high quality predictions, tuning the predictions for precision, we retained 19% of the test predictions with 100% precision. We evaluated PPI-BioBERT-x10 on 18 million PubMed abstracts and extracted 1.6 million (546507 unique PTM-PPI triplets) PTM-PPI predictions, and filter ~ 5700 (4584 unique) high confidence predictions. Of the 5700, human evaluation on a small randomly sampled subset shows that the precision drops to 33.7% despite confidence calibration and highlights the challenges of generalisability beyond the test set even with confidence calibration. We circumvent the problem by only including predictions associated with multiple papers, improving the precision to 58.8%. In this work, we highlight the benefits and challenges of deep learning-based text mining in practice, and the need for increased emphasis on confidence calibration to facilitate human curation efforts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge