Joint association and classification analysis of multi-view data
Paper and Code
Nov 20, 2018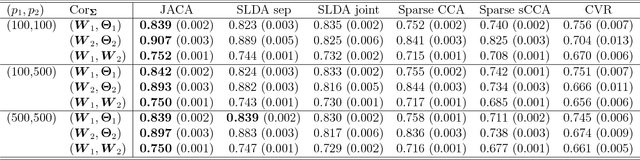
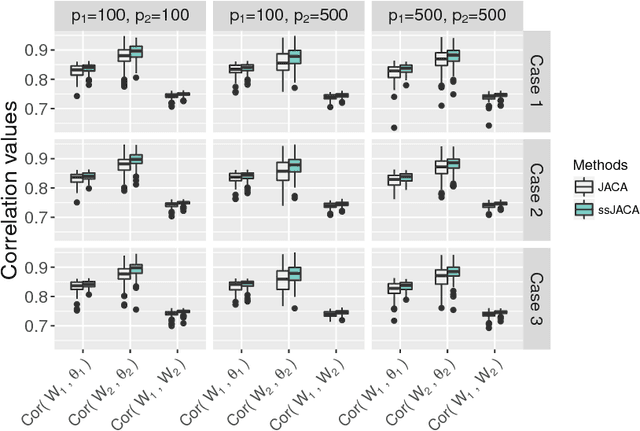
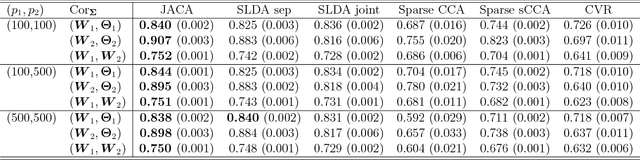
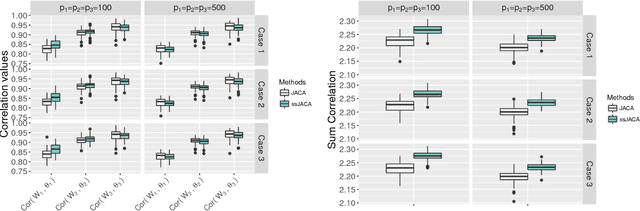
Multi-view data, that is matched sets of measurements on the same subjects, have become increasingly common with technological advances in genomics and other fields. Often, the subjects are separated into known classes, and it is of interest to find associations between the views that are related to the class membership. Existing classification methods can either be applied to each view separately, or to the concatenated matrix of all views without taking into account between-views associations. On the other hand, existing association methods can not directly incorporate class information. In this work we propose a framework for Joint Association and Classification Analysis of multi-view data (JACA). We support the methodology with theoretical guarantees for estimation consistency in high-dimensional settings, and numerical comparisons with existing methods. In addition to joint learning framework, a distinct advantage of our approach is its ability to use partial information: it can be applied both in the settings with missing class labels, and in the settings with missing subsets of views. We apply JACA to colorectal cancer data from The Cancer Genome Atlas project, and quantify the association between RNAseq and miRNA views with respect to consensus molecular subtypes of colorectal cancer.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge