Inferring Molecular Pathology and micro-RNA Transcriptome from mRNA Profiles of Cancer Biopsies through Deep Multi-Task Learning
Paper and Code
Aug 07, 2018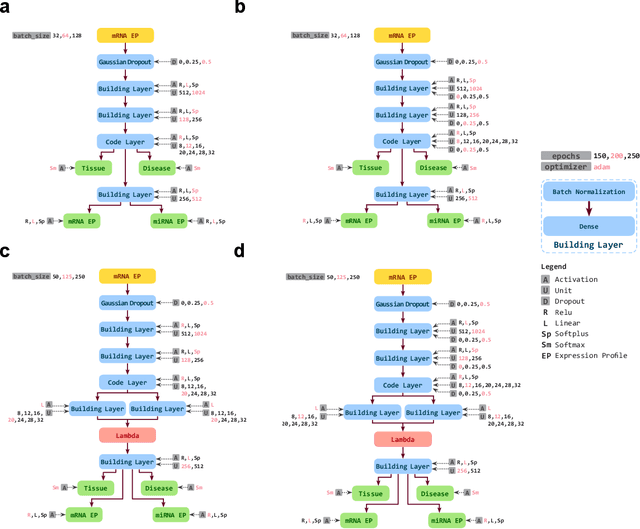
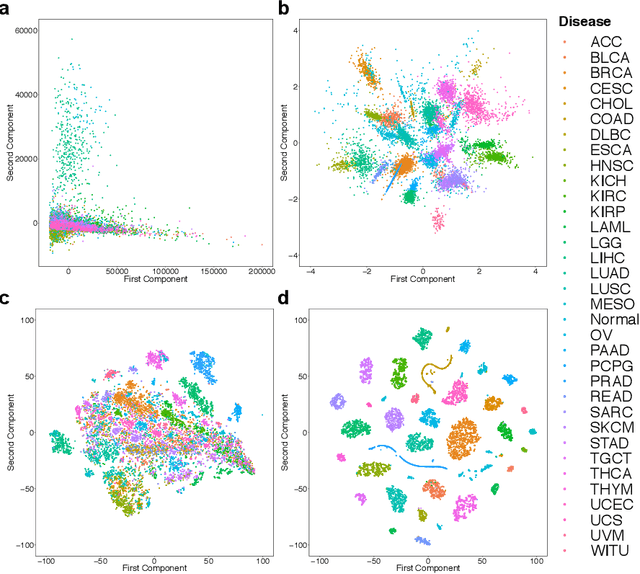
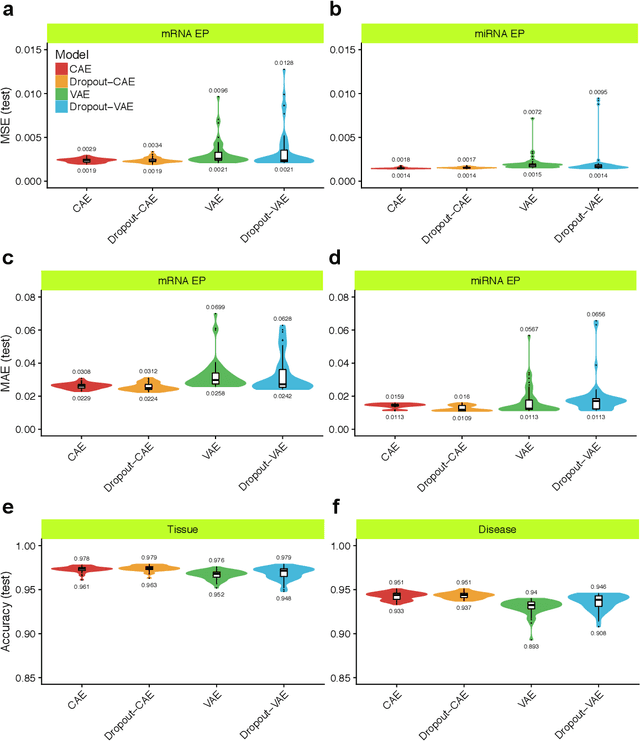
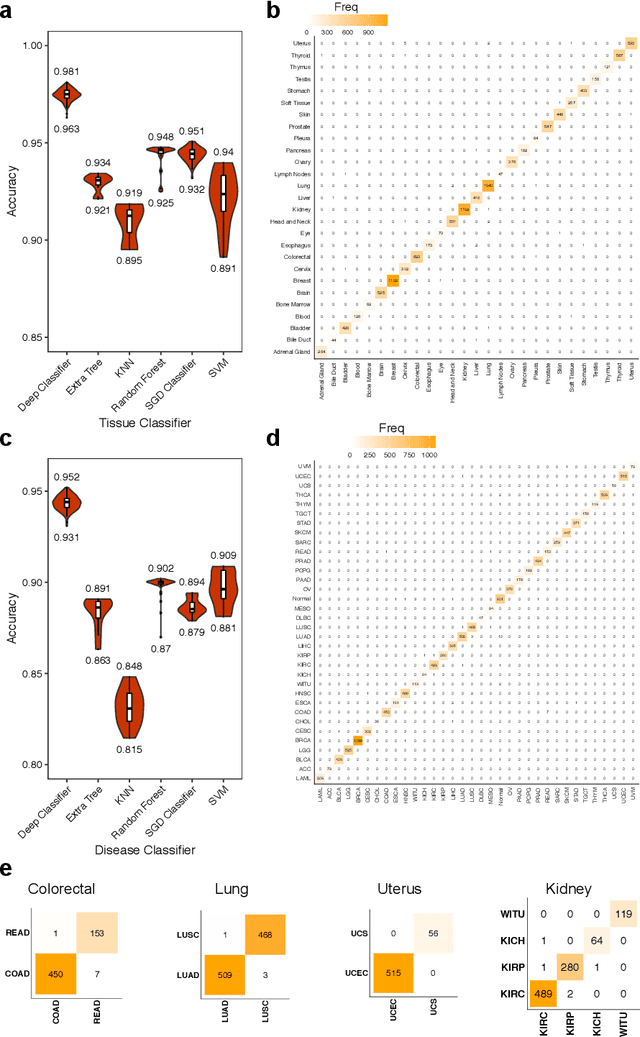
Despite great advances, molecular cancer pathology is often limited to use a small number of biomarkers rather than the whole transcriptome, partly due to the computational challenges. Here, we introduce a novel architecture of DNNs that is capable of simultaneous inference of various properties of biological samples, through multi-task and transfer learning. We employed this architecture on mRNA transcription profiles of 10787 clinical samples from 34 classes (one healthy and 33 different types of cancer) from 27 tissues. Our system significantly outperforms prior works and classical machine learning approaches in predicting tissue-of-origin, normal or disease state and cancer type of each sample. Furthermore, it can predict miRNA transcription profile of each sample, which enables performing miRNA expression research when only mRNA transcriptome data are available. We also show this system is very robust against noise and missing values. Collectively, our results highlight applications of artificial intelligence in molecular cancer pathology and oncological research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge