Inferring Disease and Gene Set Associations with Rank Coherence in Networks
Paper and Code
Feb 18, 2011
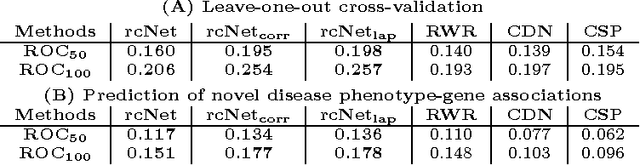

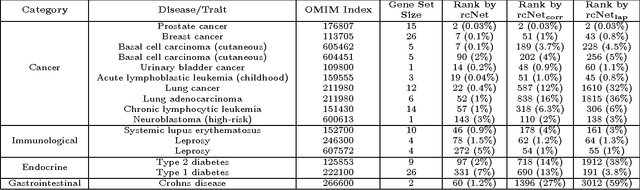
A computational challenge to validate the candidate disease genes identified in a high-throughput genomic study is to elucidate the associations between the set of candidate genes and disease phenotypes. The conventional gene set enrichment analysis often fails to reveal associations between disease phenotypes and the gene sets with a short list of poorly annotated genes, because the existing annotations of disease causative genes are incomplete. We propose a network-based computational approach called rcNet to discover the associations between gene sets and disease phenotypes. Assuming coherent associations between the genes ranked by their relevance to the query gene set, and the disease phenotypes ranked by their relevance to the hidden target disease phenotypes of the query gene set, we formulate a learning framework maximizing the rank coherence with respect to the known disease phenotype-gene associations. An efficient algorithm coupling ridge regression with label propagation, and two variants are introduced to find the optimal solution of the framework. We evaluated the rcNet algorithms and existing baseline methods with both leave-one-out cross-validation and a task of predicting recently discovered disease-gene associations in OMIM. The experiments demonstrated that the rcNet algorithms achieved the best overall rankings compared to the baselines. To further validate the reproducibility of the performance, we applied the algorithms to identify the target diseases of novel candidate disease genes obtained from recent studies of GWAS, DNA copy number variation analysis, and gene expression profiling. The algorithms ranked the target disease of the candidate genes at the top of the rank list in many cases across all the three case studies. The rcNet algorithms are available as a webtool for disease and gene set association analysis at http://compbio.cs.umn.edu/dgsa_rcNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge