Hierarchical Classification of Enzyme Promiscuity Using Positive, Unlabeled, and Hard Negative Examples
Paper and Code
Feb 18, 2020

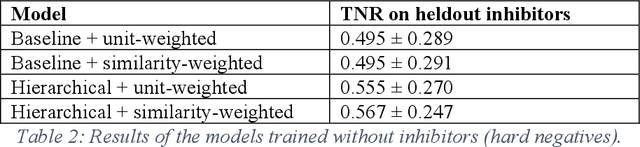
Despite significant progress in sequencing technology, there are many cellular enzymatic activities that remain unknown. We develop a new method, referred to as SUNDRY (Similarity-weighting for UNlabeled Data in a Residual HierarchY), for training enzyme-specific predictors that take as input a query substrate molecule and return whether the enzyme would act on that substrate or not. When addressing this enzyme promiscuity prediction problem, a major challenge is the lack of abundant labeled data, especially the shortage of labeled data for negative cases (enzyme-substrate pairs where the enzyme does not act to transform the substrate to a product molecule). To overcome this issue, our proposed method can learn to classify a target enzyme by sharing information from related enzymes via known tree hierarchies. Our method can also incorporate three types of data: those molecules known to be catalyzed by an enzyme (positive cases), those with unknown relationships (unlabeled cases), and molecules labeled as inhibitors for the enzyme. We refer to inhibitors as hard negative cases because they may be difficult to classify well: they bind to the enzyme, like positive cases, but are not transformed by the enzyme. Our method uses confidence scores derived from structural similarity to treat unlabeled examples as weighted negatives. We compare our proposed hierarchy-aware predictor against a baseline that cannot share information across related enzymes. Using data from the BRENDA database, we show that each of our contributions (hierarchical sharing, per-example confidence weighting of unlabeled data based on molecular similarity, and including inhibitors as hard-negative examples) contributes towards a better characterization of enzyme promiscuity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge